Epigenetic regulation of asthma and allergic disease
- PMID: 24932182
- PMCID: PMC4057652
- DOI: 10.1186/1710-1492-10-27
Epigenetic regulation of asthma and allergic disease
Abstract
Epigenetics of asthma and allergic disease is a field that has expanded greatly in the last decade. Previously thought only in terms of cell differentiation, it is now evident the epigenetics regulate many processes. With T cell activation, commitment toward an allergic phenotype is tightly regulated by DNA methylation and histone modifications at the Th2 locus control region. When normal epigenetic control is disturbed, either experimentally or by environmental exposures, Th1/Th2 balance can be affected. Epigenetic marks are not only transferred to daughter cells with cell replication but they can also be inherited through generations. In animal models, with constant environmental pressure, epigenetically determined phenotypes are amplified through generations and can last up to 2 generations after the environment is back to normal. In this review on the epigenetic regulation of asthma and allergic diseases we review basic epigenetic mechanisms and discuss the epigenetic control of Th2 cells. We then cover the transgenerational inheritance model of epigenetic traits and discuss how this could relate the amplification of asthma and allergic disease prevalence and severity through the last decades. Finally, we discuss recent epigenetic association studies for allergic phenotypes and related environmental risk factors as well as potential underlying mechanisms for these associations.
Keywords: Allergy; Amplification hypothesis; Asthma; Atopy; Epigenetic; Histone; Inheritance; Methylation; Th2; Transgenerational.
Figures

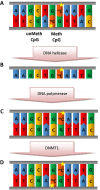
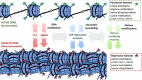

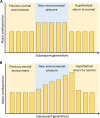

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

