Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination
- PMID: 24932489
- PMCID: PMC4059689
- DOI: 10.1371/journal.pone.0099642
Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination
Abstract
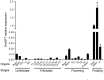
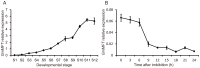
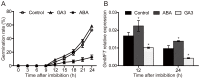
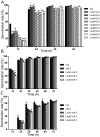
Seed germination is an important event in the life cycle of seed plants, and is controlled by complex and coordinated genetic networks. Many genes involved in the regulation of this process have been identified in different plant species so far. Recent studies in both Arabidopsis and wheat have uncovered a new role of MOTHER OF FT AND TFL1 (MFT) in seed germination. Here, we reported a homolog of MFT in soybean (GmMFT) which strongly expressed in seeds. Detailed expression analysis showed that the mRNA level of GmMFT increased with seed development but declined during seed germination. The transcription of GmMFT also responded to exogenous application of ABA and GA3. Ectopic expression of GmMFT CDS in Arabidopsis moderately inhibited seed germination. All these evidences suggest that GmMFT may be a negative regulator of seed germination.
Conflict of interest statement
Figures



Similar articles
-
Identification of cotton MOTHER OF FT AND TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination.PLoS One. 2019 Apr 19;14(4):e0215771. doi: 10.1371/journal.pone.0215771. eCollection 2019. PLoS One. 2019. PMID: 31002698 Free PMC article.
-
Identification, Functional Study, and Promoter Analysis of HbMFT1, a Homolog of MFT from Rubber Tree (Hevea brasiliensis).Int J Mol Sci. 2016 Mar 2;17(3):247. doi: 10.3390/ijms17030247. Int J Mol Sci. 2016. PMID: 26950112 Free PMC article.
-
MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis.Plant Cell. 2010 Jun;22(6):1733-48. doi: 10.1105/tpc.109.073072. Epub 2010 Jun 15. Plant Cell. 2010. PMID: 20551347 Free PMC article.
-
Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination.Plant Physiol. 2003 Sep;133(1):135-44. doi: 10.1104/pp.103.025569. Plant Physiol. 2003. PMID: 12970481 Free PMC article.
-
Transcriptome- and proteome-wide analyses of seed germination.C R Biol. 2008 Oct;331(10):815-22. doi: 10.1016/j.crvi.2008.07.023. Epub 2008 Sep 4. C R Biol. 2008. PMID: 18926496 Review.
Cited by
-
Identification of cotton MOTHER OF FT AND TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination.PLoS One. 2019 Apr 19;14(4):e0215771. doi: 10.1371/journal.pone.0215771. eCollection 2019. PLoS One. 2019. PMID: 31002698 Free PMC article.
-
KnetMiner: a comprehensive approach for supporting evidence-based gene discovery and complex trait analysis across species.Plant Biotechnol J. 2021 Aug;19(8):1670-1678. doi: 10.1111/pbi.13583. Epub 2021 Apr 5. Plant Biotechnol J. 2021. PMID: 33750020 Free PMC article.
-
Identification, evolution and expression analyses of the whole genome-wide PEBP gene family in Brassica napus L.BMC Genom Data. 2023 May 3;24(1):27. doi: 10.1186/s12863-023-01127-4. BMC Genom Data. 2023. PMID: 37138210 Free PMC article.
-
Knock-out of TERMINAL FLOWER 1 genes altered flowering time and plant architecture in Brassica napus.BMC Genet. 2020 May 19;21(1):52. doi: 10.1186/s12863-020-00857-z. BMC Genet. 2020. PMID: 32429836 Free PMC article.
-
Increasing abscisic acid levels by immunomodulation in barley grains induces precocious maturation without changing grain composition.J Exp Bot. 2016 Apr;67(9):2675-87. doi: 10.1093/jxb/erw102. Epub 2016 Mar 7. J Exp Bot. 2016. PMID: 26951372 Free PMC article.
References
-
- Weitbrecht K, Muller K, Leubner-Metzger G (2011) First off the mark: early seed germination. J Exp Bot 62: 3289–3309. - PubMed
-
- Holdsworth MJ, Bentsink L, Soppe WJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54. - PubMed
-
- Lefebvre V, North H, Frey A, Sotta B, Seo M, et al. (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45: 309–319. - PubMed
-
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, et al. (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45: 1694–1703. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

