A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement
- PMID: 24934289
- PMCID: PMC4107737
- DOI: 10.1093/brain/awu138
A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement
Abstract
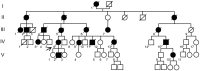
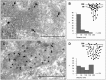
Mitochondrial DNA instability disorders are responsible for a large clinical spectrum, among which amyotrophic lateral sclerosis-like symptoms and frontotemporal dementia are extremely rare. We report a large family with a late-onset phenotype including motor neuron disease, cognitive decline resembling frontotemporal dementia, cerebellar ataxia and myopathy. In all patients, muscle biopsy showed ragged-red and cytochrome c oxidase-negative fibres with combined respiratory chain deficiency and abnormal assembly of complex V. The multiple mitochondrial DNA deletions found in skeletal muscle revealed a mitochondrial DNA instability disorder. Patient fibroblasts present with respiratory chain deficiency, mitochondrial ultrastructural alterations and fragmentation of the mitochondrial network. Interestingly, expression of matrix-targeted photoactivatable GFP showed that mitochondrial fusion was not inhibited in patient fibroblasts. Using whole-exome sequencing we identified a missense mutation (c.176C>T; p.Ser59Leu) in the CHCHD10 gene that encodes a coiled-coil helix coiled-coil helix protein, whose function is unknown. We show that CHCHD10 is a mitochondrial protein located in the intermembrane space and enriched at cristae junctions. Overexpression of a CHCHD10 mutant allele in HeLa cells led to fragmentation of the mitochondrial network and ultrastructural major abnormalities including loss, disorganization and dilatation of cristae. The observation of a frontotemporal dementia-amyotrophic lateral sclerosis phenotype in a mitochondrial disease led us to analyse CHCHD10 in a cohort of 21 families with pathologically proven frontotemporal dementia-amyotrophic lateral sclerosis. We identified the same missense p.Ser59Leu mutation in one of these families. This work opens a novel field to explore the pathogenesis of the frontotemporal dementia-amyotrophic lateral sclerosis clinical spectrum by showing that mitochondrial disease may be at the origin of some of these phenotypes.
Keywords: CHCHD10; FTD-ALS; mitochondrial DNA instability; mitochondrial disorder.
© The Author (2014). Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures







Comment in
-
Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease.Brain. 2014 Dec;137(Pt 12):e309. doi: 10.1093/brain/awu227. Epub 2014 Aug 11. Brain. 2014. PMID: 25113787 No abstract available.
-
Reply: Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease.Brain. 2014 Dec;137(Pt 12):e310. doi: 10.1093/brain/awu228. Epub 2014 Aug 11. Brain. 2014. PMID: 25113788 Free PMC article. No abstract available.
-
Reply: Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis.Brain. 2014 Dec;137(Pt 12):e312. doi: 10.1093/brain/awu267. Epub 2014 Sep 26. Brain. 2014. PMID: 25261971 Free PMC article. No abstract available.
-
Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis.Brain. 2014 Dec;137(Pt 12):e311. doi: 10.1093/brain/awu265. Epub 2014 Sep 26. Brain. 2014. PMID: 25261972 Free PMC article. No abstract available.
-
Are CHCHD10 mutations indeed associated with familial amyotrophic lateral sclerosis?Brain. 2014 Dec;137(Pt 12):e313. doi: 10.1093/brain/awu299. Epub 2014 Oct 27. Brain. 2014. PMID: 25348631 No abstract available.
-
Reply: Are CHCHD10 mutations indeed associated with familial amyotrophic lateral sclerosis?Brain. 2014 Dec;137(Pt 12):e314. doi: 10.1093/brain/awu300. Epub 2014 Oct 27. Brain. 2014. PMID: 25348633 Free PMC article. No abstract available.
-
CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis.Brain. 2015 Aug;138(Pt 8):e372. doi: 10.1093/brain/awu384. Epub 2015 Jan 8. Brain. 2015. PMID: 25576308 No abstract available.
-
Reply: CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis.Brain. 2015 Aug;138(Pt 8):e373. doi: 10.1093/brain/awu385. Epub 2015 Jan 8. Brain. 2015. PMID: 25576309 Free PMC article. No abstract available.
-
Reply: A distinct clinical phenotype in a German kindred with motor neuron disease carrying a CHCHD10 mutation.Brain. 2015 Sep;138(Pt 9):e377. doi: 10.1093/brain/awv015. Epub 2015 Feb 12. Brain. 2015. PMID: 25681413 Free PMC article. No abstract available.
-
A distinct clinical phenotype in a German kindred with motor neuron disease carrying a CHCHD10 mutation.Brain. 2015 Sep;138(Pt 9):e376. doi: 10.1093/brain/awv014. Epub 2015 Feb 12. Brain. 2015. PMID: 25681414 No abstract available.
-
Mutation analysis of CHCHD10 in different neurodegenerative diseases.Brain. 2015 Sep;138(Pt 9):e380. doi: 10.1093/brain/awv082. Epub 2015 Mar 31. Brain. 2015. PMID: 25833818 Free PMC article. No abstract available.
-
Reply: Is CHCHD10 Pro34Ser pathogenic for frontotemporal dementia and amyotrophic lateral sclerosis?Brain. 2015 Oct;138(Pt 10):e386. doi: 10.1093/brain/awv116. Epub 2015 May 7. Brain. 2015. PMID: 25953779 No abstract available.
-
Is CHCHD10 Pro34Ser pathogenic for frontotemporal dementia and amyotrophic lateral sclerosis?Brain. 2015 Oct;138(Pt 10):e385. doi: 10.1093/brain/awv115. Epub 2015 May 7. Brain. 2015. PMID: 25953780 No abstract available.
-
Screening for CHCHD10 mutations in a large cohort of sporadic ALS patients: no evidence for pathogenicity of the p.P34S variant.Brain. 2016 Feb;139(Pt 2):e8. doi: 10.1093/brain/awv218. Epub 2015 Sep 11. Brain. 2016. PMID: 26362909 Free PMC article. No abstract available.
-
Reply: High prevalence of CHCHD10 mutations in patients with frontotemporal dementia from China.Brain. 2016 Apr;139(Pt 4):e22. doi: 10.1093/brain/awv368. Epub 2015 Dec 30. Brain. 2016. PMID: 26719380 Free PMC article. No abstract available.
-
High prevalence of CHCHD10 mutation in patients with frontotemporal dementia from China.Brain. 2016 Apr;139(Pt 4):e21. doi: 10.1093/brain/awv367. Epub 2015 Dec 30. Brain. 2016. PMID: 26719383 No abstract available.
References
-
- Amati-Bonneau P, Valentino M, Reynier P, Gallardo M, Bornstein B, Boissière A, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus' phenotype. Brain. 2008;131:338–51. - PubMed
-
- Banci L, Bertini I, Ciofi-Baffoni S, Tokatlidis K. The coiled coil-helix coil-helix proteins may be redox proteins. FEBS Lett. 2009;583:1699–702. - PubMed
-
- Banci L, Bertini I, Coffi-Baffoni S, Jaiswal D, Peruzzini R, Winkelman J. Structural characterization of CHCHD5 and CHCHD7: two atypical twin CX9C proteins. J Struct Biol. 2012;180:190–200. - PubMed
-
- Bannwarth S, Figueroa A, Fragaki K, Destroismaisons L, Lacas-Gervais S, Lespinasse F, et al. The human MSH5 (MutS Homolog 5) gene localizes to mitochondria and protects the mitochondrial genome from oxidative damage. Mitochondrion. 2012;12:654–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

