A broad survey reveals substitution tolerance of residues ligating FeS clusters in [NiFe] hydrogenase
- PMID: 24934472
- PMCID: PMC4070099
- DOI: 10.1186/1471-2091-15-10
A broad survey reveals substitution tolerance of residues ligating FeS clusters in [NiFe] hydrogenase
Abstract
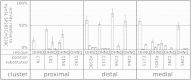
Background: In order to understand the effects of FeS cluster attachment in [NiFe] hydrogenase, we undertook a study to substitute all 12 amino acid positions normally ligating the three FeS clusters in the hydrogenase small subunit. Using the hydrogenase from Alteromonas macleodii "deep ecotype" as a model, we substituted one of four amino acids (Asp, His, Asn, Gln) at each of the 12 ligating positions because these amino acids are alternative coordinating residues in otherwise conserved-cysteine positions found in a broad survey of NiFe hydrogenase sequences. We also hoped to discover an enzyme with elevated hydrogen evolution activity relative to a previously reported "G1" (H230C/P285C) improved enzyme in which the medial FeS cluster Pro and the distal FeS cluster His were each substituted for Cys.
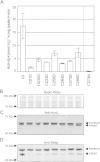
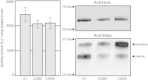
Results: Among all the substitutions screened, aspartic acid substitutions were generally well-tolerated, and examination suggests that the observed deficiency in enzyme activity may be largely due to misprocessing of the small subunit of the enzyme. Alignment of hydrogenase sequences from sequence databases revealed many rare substitutions; the five substitutions present in databases that we tested all exhibited measurable hydrogen evolution activity. Select substitutions were purified and tested, supporting the results of the screening assay. Analysis of these results confirms the importance of small subunit processing. Normalizing activity to quantity of mature small subunit, indicative of total enzyme maturation, weakly suggests an improvement over the "G1" enzyme.
Conclusions: We have comprehensively screened 48 amino acid substitutions of the hydrogenase from A. macleodii "deep ecotype", to understand non-canonical ligations of amino acids to FeS clusters and to improve hydrogen evolution activity of this class of hydrogenase. Our studies show that non-canonical ligations can be functional and also suggests a new limiting factor in the production of active enzyme.
Figures
Similar articles
-
Designed surface residue substitutions in [NiFe] hydrogenase that improve electron transfer characteristics.Int J Mol Sci. 2015 Jan 16;16(1):2020-33. doi: 10.3390/ijms16012020. Int J Mol Sci. 2015. PMID: 25603181 Free PMC article.
-
Dual organism design cycle reveals small subunit substitutions that improve [NiFe] hydrogenase hydrogen evolution.J Biol Eng. 2013 Jul 2;7(1):17. doi: 10.1186/1754-1611-7-17. J Biol Eng. 2013. PMID: 23819621 Free PMC article.
-
Genetic analysis of the Alteromonas macleodii [NiFe]-hydrogenase.FEMS Microbiol Lett. 2011 Sep;322(2):180-7. doi: 10.1111/j.1574-6968.2011.02348.x. Epub 2011 Jul 19. FEMS Microbiol Lett. 2011. PMID: 21718346
-
Structure-function relationships among the nickel-containing hydrogenases.FEMS Microbiol Rev. 1992 Feb;8(2):109-35. doi: 10.1111/j.1574-6968.1992.tb04960.x. FEMS Microbiol Rev. 1992. PMID: 1558764 Review.
-
A novel FeS cluster in Fe-only hydrogenases.Trends Biochem Sci. 2000 Mar;25(3):138-43. doi: 10.1016/s0968-0004(99)01536-4. Trends Biochem Sci. 2000. PMID: 10694885 Review.
Cited by
-
Cyanobacterial hydrogenases and hydrogen metabolism revisited: recent progress and future prospects.Int J Mol Sci. 2015 May 8;16(5):10537-61. doi: 10.3390/ijms160510537. Int J Mol Sci. 2015. PMID: 26006225 Free PMC article. Review.
-
Second and Outer Coordination Sphere Effects in Nitrogenase, Hydrogenase, Formate Dehydrogenase, and CO Dehydrogenase.Chem Rev. 2022 Jul 27;122(14):11900-11973. doi: 10.1021/acs.chemrev.1c00914. Epub 2022 Jul 18. Chem Rev. 2022. PMID: 35849738 Free PMC article. Review.
-
Influence of the [4Fe-4S] cluster coordinating cysteines on active site maturation and catalytic properties of C. reinhardtii [FeFe]-hydrogenase.Chem Sci. 2017 Dec 1;8(12):8127-8137. doi: 10.1039/c7sc03444j. Epub 2017 Oct 9. Chem Sci. 2017. PMID: 29568461 Free PMC article.
-
From protein engineering to artificial enzymes - biological and biomimetic approaches towards sustainable hydrogen production.Sustain Energy Fuels. 2018 Apr 1;2(4):724-750. doi: 10.1039/c7se00582b. Epub 2018 Feb 6. Sustain Energy Fuels. 2018. PMID: 31497651 Free PMC article. Review.
-
Designed surface residue substitutions in [NiFe] hydrogenase that improve electron transfer characteristics.Int J Mol Sci. 2015 Jan 16;16(1):2020-33. doi: 10.3390/ijms16012020. Int J Mol Sci. 2015. PMID: 25603181 Free PMC article.
References
-
- Gallon JR. The oxygen sensitivity of nitrogenase: a problem for biochemists and micro-organisms. Trends Biochem Sci. 1981;6:19–23.
-
- Bingham AS, Smith PR, Swartz JR. Evolution of an [FeFe] hydrogenase with decreased oxygen sensitivity. Int J Hydrog Energy. 2012;37:2965–2976. doi: 10.1016/j.ijhydene.2011.02.048. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

