Cytotoxicity of the Vibrio vulnificus MARTX toxin effector DUF5 is linked to the C2A subdomain
- PMID: 24935440
- PMCID: PMC4177264
- DOI: 10.1002/prot.24628
Cytotoxicity of the Vibrio vulnificus MARTX toxin effector DUF5 is linked to the C2A subdomain
Abstract
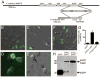
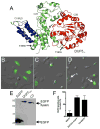
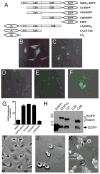
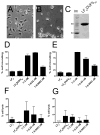
The multifunctional-autoprocessing repeats-in-toxin (MARTX) toxins are bacterial protein toxins that serve as delivery platforms for cytotoxic effector domains. The domain of unknown function in position 5 (DUF5) effector domain is present in at least six different species' MARTX toxins and as a hypothetical protein in Photorhabdus spp. Its presence increases the potency of the Vibrio vulnificus MARTX toxin in mouse virulence studies, indicating DUF5 directly contributes to pathogenesis. In this work, DUF5 is shown to be cytotoxic when transiently expressed in HeLa cells. DUF5 localized to the plasma membrane dependent upon its C1 domain and the cells become rounded dependent upon its C2 domain. Both full-length DUF5 and the C2 domain caused growth inhibition when expressed in Saccharomyces cerevisiae. A structural model of DUF5 was generated based on the structure of Pasteurella multocida toxin facilitating localization of the cytotoxic activity to a 186 amino acid subdomain termed C2A. Within this subdomain, an alanine scanning mutagenesis revealed aspartate-3721 and arginine-3841 as residues critical for cytotoxicity. These residues were also essential for HeLa cell intoxication when purified DUF5 fused to anthrax toxin lethal factor was delivered cytosolically. Thermal shift experiments indicated that these conserved residues are important to maintain protein structure, rather than for catalysis. The Aeromonas hydrophila MARTX toxin DUF5(Ah) domain was also cytotoxic, while the weakly conserved C1-C2 domains from P. multocida toxin were not. Overall, this study is the first demonstration that DUF5 as found in MARTX toxins has cytotoxic activity that depends on conserved residues in the C2A subdomain.
Keywords: MARTX; Vibrio vulnificus; cytotoxin; effector domains; yeast screen.
© 2014 Wiley Periodicals, Inc.
Figures





References
-
- Satchell KJ. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol. 2011;65:71–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

