The determination of tRNALeu recognition nucleotides for Escherichia coli L/F transferase
- PMID: 24935875
- PMCID: PMC4105747
- DOI: 10.1261/rna.044529.114
The determination of tRNALeu recognition nucleotides for Escherichia coli L/F transferase
Abstract
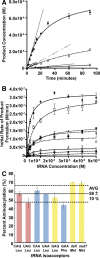
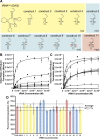
Escherichia coli leucyl/phenylalanyl-tRNA protein transferase catalyzes the tRNA-dependent post-translational addition of amino acids onto the N-terminus of a protein polypeptide substrate. Based on biochemical and structural studies, the current tRNA recognition model by L/F transferase involves the identity of the 3' aminoacyl adenosine and the sequence-independent docking of the D-stem of an aminoacyl-tRNA to the positively charged cluster on L/F transferase. However, this model does not explain the isoacceptor preference observed 40 yr ago. Using in vitro-transcribed tRNA and quantitative MALDI-ToF MS enzyme activity assays, we have confirmed that, indeed, there is a strong preference for the most abundant leucyl-tRNA, tRNA(Leu) (anticodon 5'-CAG-3') isoacceptor for L/F transferase activity. We further investigate the molecular mechanism for this preference using hybrid tRNA constructs. We identified two independent sequence elements in the acceptor stem of tRNA(Leu) (CAG)-a G₃:C₇₀ base pair and a set of 4 nt (C₇₂, A₄:U₆₉, C₆₈)-that are important for the optimal binding and catalysis by L/F transferase. This maps a more specific, sequence-dependent tRNA recognition model of L/F transferase than previously proposed.
Keywords: L/F transferase; N-end rule; aminoacyl-tRNA protein transferase; isoacceptors; quantitative mass spectrometry; tRNA recognition.
© 2014 Fung et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
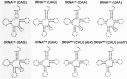
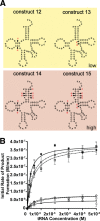
 ), 13 (⋄), 14 (⊗), and 15 (⊠). Errors represented are the standard deviation of three independent experiments.
), 13 (⋄), 14 (⊗), and 15 (⊠). Errors represented are the standard deviation of three independent experiments.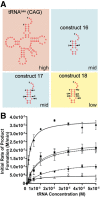

Similar articles
-
Perspectives and Insights into the Competition for Aminoacyl-tRNAs between the Translational Machinery and for tRNA Dependent Non-Ribosomal Peptide Bond Formation.Life (Basel). 2015 Dec 31;6(1):2. doi: 10.3390/life6010002. Life (Basel). 2015. PMID: 26729173 Free PMC article. Review.
-
Aminoacyl-tRNA recognition by the leucyl/phenylalanyl-tRNA-protein transferase.J Biol Chem. 1996 Sep 13;271(37):22901-7. doi: 10.1074/jbc.271.37.22901. J Biol Chem. 1996. PMID: 8798470
-
Probing the leucyl/phenylalanyl tRNA protein transferase active site with tRNA substrate analogues.Protein Pept Lett. 2014 Jul;21(7):603-14. doi: 10.2174/0929866521666140212110639. Protein Pept Lett. 2014. PMID: 24521222
-
Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps.Nucleic Acids Res. 2008 May;36(8):2728-38. doi: 10.1093/nar/gkn028. Epub 2008 Mar 26. Nucleic Acids Res. 2008. PMID: 18367476 Free PMC article.
-
The molecular basis for the post-translational addition of amino acids by L/F transferase in the N-end rule pathway.Curr Protein Pept Sci. 2015;16(2):163-80. Curr Protein Pept Sci. 2015. PMID: 25692952 Review.
Cited by
-
ATE1-Mediated Post-Translational Arginylation Is an Essential Regulator of Eukaryotic Cellular Homeostasis.ACS Chem Biol. 2020 Dec 18;15(12):3073-3085. doi: 10.1021/acschembio.0c00677. Epub 2020 Nov 23. ACS Chem Biol. 2020. PMID: 33228359 Free PMC article.
-
A [3Fe-4S] cluster and tRNA-dependent aminoacyltransferase BlsK in the biosynthesis of Blasticidin S.Proc Natl Acad Sci U S A. 2021 Jul 27;118(30):e2102318118. doi: 10.1073/pnas.2102318118. Proc Natl Acad Sci U S A. 2021. PMID: 34282016 Free PMC article.
-
Structure and tRNA Specificity of MibB, a Lantibiotic Dehydratase from Actinobacteria Involved in NAI-107 Biosynthesis.Cell Chem Biol. 2016 Mar 17;23(3):370-380. doi: 10.1016/j.chembiol.2015.11.017. Epub 2016 Feb 11. Cell Chem Biol. 2016. PMID: 26877024 Free PMC article.
-
Perspectives and Insights into the Competition for Aminoacyl-tRNAs between the Translational Machinery and for tRNA Dependent Non-Ribosomal Peptide Bond Formation.Life (Basel). 2015 Dec 31;6(1):2. doi: 10.3390/life6010002. Life (Basel). 2015. PMID: 26729173 Free PMC article. Review.
-
tRNAArg-Derived Fragments Can Serve as Arginine Donors for Protein Arginylation.Cell Chem Biol. 2020 Jul 16;27(7):839-849.e4. doi: 10.1016/j.chembiol.2020.05.013. Epub 2020 Jun 16. Cell Chem Biol. 2020. PMID: 32553119 Free PMC article.
References
-
- Abramochkin G, Shrader TE 1995. The leucyl/phenylalanyl-tRNA-protein transferase. Overexpression and characterization of substrate recognition, domain structure, and secondary structure. J Biol Chem 270: 20621–20628 - PubMed
-
- Abramochkin G, Shrader TE 1996. Aminoacyl-tRNA recognition by the leucyl/phenylalanyl-tRNA-protein transferase. J Biol Chem 271: 22901–22907 - PubMed
-
- Andersen C, Wiborg O 1994. Escherichia coli elongation-factor-Tu mutants with decreased affinity for aminoacyl-tRNA. Eur J Biochem 220: 739–744 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
