Integration of the hybrid adenoretroviral vector AdLTR-luc involves both MoMLV elements flanking the transgene
- PMID: 24936143
- PMCID: PMC4057490
- DOI: 10.7150/ijms.9084
Integration of the hybrid adenoretroviral vector AdLTR-luc involves both MoMLV elements flanking the transgene
Abstract
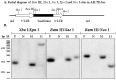
Vector delivery is still a bottleneck for gene therapy. To overcome some disadvantages of adenoviral and retroviral vectors, we developed a hybrid vector. This hybrid vector, AdLTR-luc, was created by adding two elements from Moloney murine leukemia virus (MoMLV) flanking the luciferase cDNA into an E1/E3-deleted, replication deficient serotype 5 adenovirus vector (Zheng et al., Nature Biotechnol, 2000), and demonstrated that the MoMLV element upstream of the luciferase cDNA was broken during the integration event. The purpose of the current study was to determine if the MoMLV element downstream of the luciferase cDNA was also broken when integration occurred. We used the same A5 cell clones (#10 and 11) from the earlier the paper along with restriction endonuclease digestions, plus Southern hybridization, and PCR. Southern hybridization indicated that the luciferase cDNA was intact in the cloned cells. Results from Xho I and Sal I digestions showed that integration occurred in cloned cells. Southern hybridizations after Nco I digestion suggested that there was a break in both MoMLV elements, upstream and downstream of the luciferase cDNA. After DNA digestion with Not I, hybridization analyses indicated that the MoMLV upstream element was broken during integration. Digestion of genomic DNA with either Xba I/Kpn I, Bam HI/Sac I, or Bam HI/Nco I demonstrated that the MoMLV downstream element was also broken during integration. A PCR assay was unable to amplify the junctional region between the downstream MoMLV element and the adenoviral E2B gene, consistent with a break in that element. Although AdLTR-luc integration is atypical (Zheng et al., Nature Biotechnol, 2000), the present results suggest that both MoMLV elements have important roles in this event.
Keywords: Hybrid vector; LTR; adenoretrovirus; gene therapy; integration.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Bilbao G, Feng M, Rancourt C, Jackson WH Jr, Curiel DT. Adenovi-ral/retroviral vector chimeras: a novel strategy to achieve high-efficiency stable transduction in vivo. FASEB J. 1997;11:624–34. - PubMed
-
- Feng M, Jackson WH Jr Goldman CK, Rancourt C, Wang M, Dusing SK, Siegal G, Curiel DT. Stable in vivo gene transduction via a novel adenoviral/retroviral chimeric vector. Nat Biotechnol. 1997;15:866–70. - PubMed
-
- Zheng C, Baum BJ, Iadarola MJ, O'Connell BC. Geneomic integration and gene expression by a modified adenoviral vector. Nature Biotech. 2000;18:176–180. - PubMed
-
- Dales S, Chardonnet Y. Early events in the interaction of adenoviruses with HeLa cells. IV. Association with microtubules and the nuclear pore complex during vectorial movement of the inoculum. Virology. 1973;56:465–483. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

