Up to Species-level Community Analysis of Human Gut Microbiota by 16S rRNA Amplicon Pyrosequencing
- PMID: 24936364
- PMCID: PMC4034321
- DOI: 10.12938/bmfh.32.69
Up to Species-level Community Analysis of Human Gut Microbiota by 16S rRNA Amplicon Pyrosequencing
Abstract
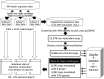
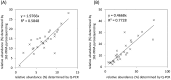
Pyrosequencing-based 16S rRNA profiling has become a common powerful tool to obtain the community structure of gastrointestinal tract microbiota, but it is still hard to process the massive amount of sequence data into microbial composition data, especially at the species level. Here we propose a new approach in combining the quantitative insights into microbial ecology (QIIME), Mothur and ribosomal database project (RDP) programs to efficiently process 454 pyrosequence data to bacterial composition data up to the species level. It was demonstrated to precisely convert batch sequence data of 16S rRNA V6-V8 amplicons obtained from adult Singaporean fecal samples to taxonomically annotated biota data.
Keywords: 16S rRNA gene; human gut microbiota; pyrosequence.
Figures


References
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336 - PMC - PubMed
-
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541 - PMC - PubMed
-
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486: 222–227 - PMC - PubMed
-
- Nakayama J. 2010. Pyrosequence-based 16S rRNA profiling of gastro-intestinal microbiota. Biosci Microflora 29: 83–96
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
