Efficient genetic manipulation of the NOD-Rag1-/-IL2RgammaC-null mouse by combining in vitro fertilization and CRISPR/Cas9 technology
- PMID: 24936832
- PMCID: PMC4894429
- DOI: 10.1038/srep05290
Efficient genetic manipulation of the NOD-Rag1-/-IL2RgammaC-null mouse by combining in vitro fertilization and CRISPR/Cas9 technology
Abstract
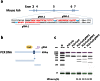
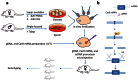
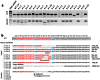
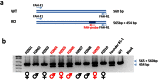
Humanized mouse models have become increasingly important and widely used in modeling human diseases in biomedical research. Immunodeficient mice such as NOD-Rag1-/-IL2RgammaC-null (NRG) or NOD-SCID-IL2RgammaC-null (NSG) mice are critical for efficient engraftment of human cells or tissues. However, their genetic modification remains challenging due to a lack of embryonic stem cells and difficulty in the collection of timed embryos after superovulation. Here, we report the generation of gene knockout NRG mice by combining in vitro fertilization (IVF) and CRISPR/Cas9 technology. Sufficient numbers of fertilized embryos were produced through IVF, and a high rate of Fah gene targeting was achieved with microinjection of Cas9 mRNA, gRNA and single strand oligonucleotide DNA (ssDNA) into the embryos. The technology paves the way to construct NRG or NSG mutant mice to facilitate new humanized mouse models. The technology can also be readily adapted to introduce mutations in other species such as swine and non-human primates.
Conflict of interest statement
Yes, there is potential Competing Interest. D. Cowley is employed by, has equity ownership in and serves on the board of directors of TransViragen, the company which has been contracted by UNC-Chapel Hill to manage its Animal Models Core Facility. No other authors have any competing financial interests.
Figures
References
-
- Takebe T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

