Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes
- PMID: 24938783
- PMCID: PMC4084482
- DOI: 10.1073/pnas.1320298111
Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes
Abstract
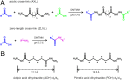
The study of proteins and protein complexes using chemical cross-linking followed by the MS identification of the cross-linked peptides has found increasingly widespread use in recent years. Thus far, such analyses have used almost exclusively homobifunctional, amine-reactive cross-linking reagents. Here we report the development and application of an orthogonal cross-linking chemistry specific for carboxyl groups. Chemical cross-linking of acidic residues is achieved using homobifunctional dihydrazides as cross-linking reagents and a coupling chemistry at neutral pH that is compatible with the structural integrity of most protein complexes. In addition to cross-links formed through insertion of the dihydrazides with different spacer lengths, zero-length cross-link products are also obtained, thereby providing additional structural information. We demonstrate the application of the reaction and the MS identification of the resulting cross-linked peptides for the chaperonin TRiC/CCT and the 26S proteasome. The results indicate that the targeting of acidic residues for cross-linking provides distance restraints that are complementary and orthogonal to those obtained from lysine cross-linking, thereby expanding the yield of structural information that can be obtained from cross-linking studies and used in hybrid modeling approaches.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Serpa JJ, et al. Mass spectrometry-based structural proteomics. Eur J Mass Spectrom (Chichester, Eng) 2012;18(2):251–267. - PubMed
-
- Walzthoeni T, Leitner A, Stengel F, Aebersold R. Mass spectrometry supported determination of protein complex structure. Curr Opin Struct Biol. 2013;23(2):252–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

