Genome-wide association analysis demonstrates the highly polygenic character of age-related hearing impairment
- PMID: 24939585
- PMCID: PMC4266741
- DOI: 10.1038/ejhg.2014.56
Genome-wide association analysis demonstrates the highly polygenic character of age-related hearing impairment
Abstract
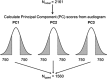
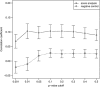
We performed a genome-wide association study (GWAS) to identify the genes responsible for age-related hearing impairment (ARHI), the most common form of hearing impairment in the elderly. Analysis of common variants, with and without adjustment for stratification and environmental covariates, rare variants and interactions, as well as gene-set enrichment analysis, showed no variants with genome-wide significance. No evidence for replication of any previously reported genes was found. A study of the genetic architecture indicates for the first time that ARHI is highly polygenic in nature, with probably no major genes involved. The phenotype depends on the aggregated effect of a large number of SNPs, of which the individual effects are undetectable in a modestly powered GWAS. We estimated that 22% of the variance in our data set can be explained by the collective effect of all genotyped SNPs. A score analysis showed a modest enrichment in causative SNPs among the SNPs with a P-value below 0.01.
Figures

References
-
- Gates GA, Mills JH. Presbycusis. Lancet. 2005;366:1111–1120. - PubMed
-
- Huang Q, Tang J. Age-related hearing loss or presbycusis. Eur Arch Otorhinolaryngol. 2010;267:1179–1191. - PubMed
-
- Girotto G, Pirastu N, Sorice R, et al. Hearing function and thresholds: a genome-wide association study in European isolated populations identifies new loci and pathways. J Med Genet. 2011;48:369–374. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

