Linking genotypes database with locus-specific database and genotype-phenotype correlation in phenylketonuria
- PMID: 24939588
- PMCID: PMC4326710
- DOI: 10.1038/ejhg.2014.114
Linking genotypes database with locus-specific database and genotype-phenotype correlation in phenylketonuria
Abstract
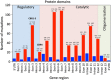
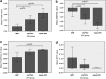
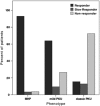
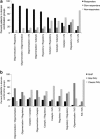
The wide range of metabolic phenotypes in phenylketonuria is due to a large number of variants causing variable impairment in phenylalanine hydroxylase function. A total of 834 phenylalanine hydroxylase gene variants from the locus-specific database PAHvdb and genotypes of 4181 phenylketonuria patients from the BIOPKU database were characterized using FoldX, SIFT Blink, Polyphen-2 and SNPs3D algorithms. Obtained data was correlated with residual enzyme activity, patients' phenotype and tetrahydrobiopterin responsiveness. A descriptive analysis of both databases was compiled and an interactive viewer in PAHvdb database was implemented for structure visualization of missense variants. We found a quantitative relationship between phenylalanine hydroxylase protein stability and enzyme activity (r(s) = 0.479), between protein stability and allelic phenotype (r(s) = -0.458), as well as between enzyme activity and allelic phenotype (r(s) = 0.799). Enzyme stability algorithms (FoldX and SNPs3D), allelic phenotype and enzyme activity were most powerful to predict patients' phenotype and tetrahydrobiopterin response. Phenotype prediction was most accurate in deleterious genotypes (≈ 100%), followed by homozygous (92.9%), hemizygous (94.8%), and compound heterozygous genotypes (77.9%), while tetrahydrobiopterin response was correctly predicted in 71.0% of all cases. To our knowledge this is the largest study using algorithms for the prediction of patients' phenotype and tetrahydrobiopterin responsiveness in phenylketonuria patients, using data from the locus-specific and genotypes database.
Figures
References
-
- Blau N, Van Spronsen FJ, Levy HL. Phenylketonuria. Lancet. 2010;376:1417–1427. - PubMed
-
- Koch R, Azen C, Friedman EG, Williamson ML. Paired comparisons between early treated PKU children and their matched sibling controls on intelligence and school achievement test results at eight years of age. J Inherit Metab Dis. 1984;7:86–90. - PubMed
-
- Waisbren SE, Noel K, Fahrbach K, et al. Phenylalanine blood levels and clinical outcomes in phenylketonuria: a systematic literature review and meta-analysis. Mol Genet Metab. 2007;92:63–70. - PubMed
-
- Pey AL, Desviat LR, Gamez A, Ugarte M, Perez B. Phenylketonuria: genotype-phenotype correlations based on expression analysis of structural and functional mutations in PAH. Hum Mutat. 2003;21:370–378. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

