Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life
- PMID: 24939692
- PMCID: PMC11113594
- DOI: 10.1007/s00018-014-1661-9
Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life
Abstract
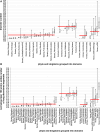
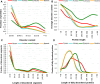
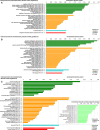
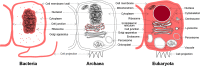
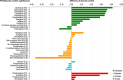
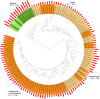
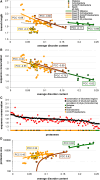
Recent years witnessed increased interest in intrinsically disordered proteins and regions. These proteins and regions are abundant and possess unique structural features and a broad functional repertoire that complements ordered proteins. However, modern studies on the abundance and functions of intrinsically disordered proteins and regions are relatively limited in size and scope of their analysis. To fill this gap, we performed a broad and detailed computational analysis of over 6 million proteins from 59 archaea, 471 bacterial, 110 eukaryotic and 325 viral proteomes. We used arguably more accurate consensus-based disorder predictions, and for the first time comprehensively characterized intrinsic disorder at proteomic and protein levels from all significant perspectives, including abundance, cellular localization, functional roles, evolution, and impact on structural coverage. We show that intrinsic disorder is more abundant and has a unique profile in eukaryotes. We map disorder into archaea, bacterial and eukaryotic cells, and demonstrate that it is preferentially located in some cellular compartments. Functional analysis that considers over 1,200 annotations shows that certain functions are exclusively implemented by intrinsically disordered proteins and regions, and that some of them are specific to certain domains of life. We reveal that disordered regions are often targets for various post-translational modifications, but primarily in the eukaryotes and viruses. Using a phylogenetic tree for 14 eukaryotic and 112 bacterial species, we analyzed relations between disorder, sequence conservation and evolutionary speed. We provide a complete analysis that clearly shows that intrinsic disorder is exceptionally and uniquely abundant in each domain of life.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

