Primary identification, biochemical characterization, and immunologic properties of the allergenic pollen cyclophilin cat R 1
- PMID: 24939849
- PMCID: PMC4118102
- DOI: 10.1074/jbc.M114.559971
Primary identification, biochemical characterization, and immunologic properties of the allergenic pollen cyclophilin cat R 1
Abstract
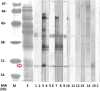
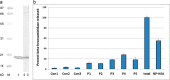
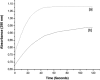
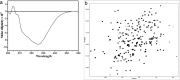
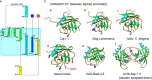
Cyclophilin (Cyp) allergens are considered pan-allergens due to frequently reported cross-reactivity. In addition to well studied fungal Cyps, a number of plant Cyps were identified as allergens (e.g. Bet v 7 from birch pollen, Cat r 1 from periwinkle pollen). However, there are conflicting data regarding their antigenic/allergenic cross-reactivity, with no plant Cyp allergen structures available for comparison. Because amino acid residues are fairly conserved between plant and fungal Cyps, it is particularly interesting to check whether they can cross-react. Cat r 1 was identified by immunoblotting using allergic patients' sera followed by N-terminal sequencing. Cat r 1 (∼ 91% sequence identity to Bet v 7) was cloned from a cDNA library and expressed in Escherichia coli. Recombinant Cat r 1 was utilized to confirm peptidyl-prolyl cis-trans-isomerase (PPIase) activity by a PPIase assay and the allergenic property by an IgE-specific immunoblotting and rat basophil leukemia cell (RBL-SX38) mediator release assay. Inhibition-ELISA showed cross-reactive binding of serum IgE from Cat r 1-allergic individuals to fungal allergenic Cyps Asp f 11 and Mala s 6. The molecular structure of Cat r 1 was determined by NMR spectroscopy. The antigenic surface was examined in relation to its plant, animal, and fungal homologues. The structure revealed a typical cyclophilin fold consisting of a compact β-barrel made up of seven anti-parallel β-strands along with two surrounding α-helices. This is the first structure of an allergenic plant Cyp revealing high conservation of the antigenic surface particularly near the PPIase active site, which supports the pronounced cross-reactivity among Cyps from various sources.
Keywords: Allergen; Antibody; Antigen; Cloning; Cross-reactivity; Cyclophilin; Nuclear Magnetic Resonance (NMR); Protein Structure.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Schreiber S. L. (1991) Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251, 283–287 - PubMed
-
- Marín-Menéndez A., Monaghan P., Bell A. (2012) A family of cyclophilin-like molecular chaperones in Plasmodium falciparum. Mol Biochem. Parasitol. 184, 44–47 - PubMed
-
- Limacher A., Kloer D. P., Flückiger S., Folkers G., Crameri R., Scapozza L. (2006) The crystal structure of Aspergillus fumigatus cyclophilin reveals 3D domain swapping of a central element. Structure 14, 185–195 - PubMed
-
- Crameri R. (1999) Epidemiology and molecular basis of the involvement of Aspergillus fumigatus in allergic diseases. Contrib. Microbiol. 2, 44–56 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

