The elusive evidence for chromothripsis
- PMID: 24939897
- PMCID: PMC4117757
- DOI: 10.1093/nar/gku525
The elusive evidence for chromothripsis
Abstract
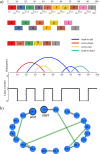
The chromothripsis hypothesis suggests an extraordinary one-step catastrophic genomic event allowing a chromosome to 'shatter into many pieces' and reassemble into a functioning chromosome. Recent efforts have aimed to detect chromothripsis by looking for a genomic signature, characterized by a large number of breakpoints (50-250), but a limited number of oscillating copy number states (2-3) confined to a few chromosomes. The chromothripsis phenomenon has become widely reported in different cancers, but using inconsistent and sometimes relaxed criteria for determining rearrangements occur simultaneously rather than progressively. We revisit the original simulation approach and show that the signature is not clearly exceptional, and can be explained using only progressive rearrangements. For example, 3.9% of progressively simulated chromosomes with 50-55 breakpoints were dominated by two or three copy number states. In addition, by adjusting the parameters of the simulation, the proposed footprint appears more frequently. Lastly, we provide an algorithm to find a sequence of progressive rearrangements that explains all observed breakpoints from a proposed chromothripsis chromosome. Thus, the proposed signature cannot be considered a sufficient proof for this extraordinary hypothesis. Great caution should be exercised when labeling complex rearrangements as chromothripsis from genome hybridization and sequencing experiments.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Molenaar J.J., Koster J., Zwijnenburg D.A., van Sluis P., Valentijn L.J., van der Ploeg I., Hamdi M., van Nes J., Westerman B.A., van Arkel J., et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

