Gene expression correlations in human cancer cell lines define molecular interaction networks for epithelial phenotype
- PMID: 24940735
- PMCID: PMC4062414
- DOI: 10.1371/journal.pone.0099269
Gene expression correlations in human cancer cell lines define molecular interaction networks for epithelial phenotype
Abstract
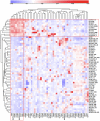
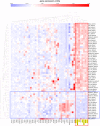
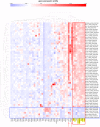
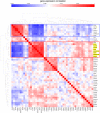
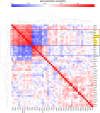
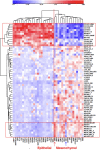
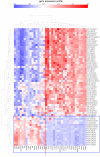
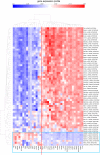
Using gene expression data to enhance our knowledge of control networks relevant to cancer biology and therapy is a challenging but urgent task. Based on the premise that genes that are expressed together in a variety of cell types are likely to functions together, we derived mutually correlated genes that function together in various processes in epithelial-like tumor cells. Expression-correlated genes were derived from data for the NCI-60 human tumor cell lines, as well as data from the Broad Institute's CCLE cell lines. NCI-60 cell lines that selectively expressed a mutually correlated subset of tight junction genes served as a signature for epithelial-like cancer cells. Those signature cell lines served as a seed to derive other correlated genes, many of which had various other epithelial-related functions. Literature survey yielded molecular interaction and function information about those genes, from which molecular interaction maps were assembled. Many of the genes had epithelial functions unrelated to tight junctions, demonstrating that new function categories were elicited. The most highly correlated genes were implicated in the following epithelial functions: interactions at tight junctions (CLDN7, CLDN4, CLDN3, MARVELD3, MARVELD2, TJP3, CGN, CRB3, LLGL2, EPCAM, LNX1); interactions at adherens junctions (CDH1, ADAP1, CAMSAP3); interactions at desmosomes (PPL, PKP3, JUP); transcription regulation of cell-cell junction complexes (GRHL1 and 2); epithelial RNA splicing regulators (ESRP1 and 2); epithelial vesicle traffic (RAB25, EPN3, GRHL2, EHF, ADAP1, MYO5B); epithelial Ca(+2) signaling (ATP2C2, S100A14, BSPRY); terminal differentiation of epithelial cells (OVOL1 and 2, ST14, PRSS8, SPINT1 and 2); maintenance of apico-basal polarity (RAB25, LLGL2, EPN3). The findings provide a foundation for future studies to elucidate the functions of regulatory networks specific to epithelial-like cancer cells and to probe for anti-cancer drug targets.
Conflict of interest statement
Figures






References
-
- Wang Q, Margolis B (2007) Apical junctional complexes and cell polarity. Kidney Int 72: 1448–1458. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

