Whole brain expression of bipolar disorder associated genes: structural and genetic analyses
- PMID: 24941232
- PMCID: PMC4062532
- DOI: 10.1371/journal.pone.0100204
Whole brain expression of bipolar disorder associated genes: structural and genetic analyses
Abstract
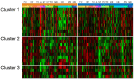
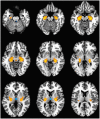
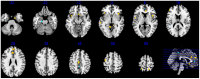
Studies of bipolar disorder (BD) suggest a genetic basis of the illness that alters brain function and morphology. In recent years, a number of genetic variants associated with BD have been identified. However, little is known about the associated genes, or brain circuits that rely upon their function. Using an anatomically comprehensive survey of the human transcriptome (The Allen Brain Atlas), we mapped the expression of 58 genes with suspected involvement in BD based upon their relationship to SNPs identified in genome wide association studies (GWAS). We then conducted a meta-analysis of structural MRI studies to identify brain regions that are abnormal in BD. Of 58 BD associated genes, 22 had anatomically distinct expression patterns that could be categorized into one of three clusters (C1-C3). Brain regions with the highest and lowest expression of these genes did not overlap strongly with anatomical sites identified as abnormal by structural MRI except in the parahippocampal gyrus, the inferior/superior temporal gyrus and the cerebellar vermis, regions where overlap was significant. Using the 22 genes in C1-C3 as reference points, additional genes with correlated expression patterns were identified and organized into sets based on similarity. Further analysis revealed that five of these gene sets were significantly associated with BD, suggesting that anatomical expression profile is correlated with genetic susceptibility to BD, particularly for genes in C2. Our data suggest that expression profiles of BD-associated genes do not explain the majority of structural abnormalities observed in BD, but may be useful in identifying new candidate genes. Our results highlight the complex neuroanatomical basis of BD, and reinforce illness models that emphasize impaired brain connectivity.
Conflict of interest statement
Figures



References
-
- McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, et al. (2003) The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry 60: 497–502. - PubMed
-
- Rimol LM, Hartberg CB, Nesvag R, Fennema-Notestine C, Hagler DJ Jr, et al. (2010) Cortical thickness and subcortical volumes in schizophrenia and bipolar disorder. Biol Psychiatry 68: 41–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

