Phosphoproteome profiling of the macrophage response to different toll-like receptor ligands identifies differences in global phosphorylation dynamics
- PMID: 24941444
- PMCID: PMC4227906
- DOI: 10.1021/pr5002466
Phosphoproteome profiling of the macrophage response to different toll-like receptor ligands identifies differences in global phosphorylation dynamics
Abstract
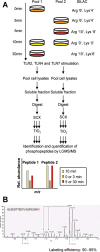
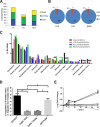
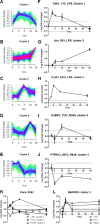
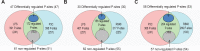
Toll-like receptors (TLRs) are among the first sensors that detect infection and drive immune response. Macrophages encountering a pathogen are usually stimulated not by one TLR, but by a combination of TLRs engaged by distinct microbe ligands. To understand the integrated signaling under complex conditions, we investigated the differences in the phosphoprotein signaling cascades triggered by TLR2, TLR4, and TLR7 ligands using a single responding cell population. We performed a global, quantitative, early poststimulation kinetic analysis of the mouse macrophage phosphoproteome using stable isotope labeling with amino acids coupled to phosphopeptide enrichment and high-resolution mass spectrometry. For each TLR ligand, we found marked elevation of phosphorylation of cytoskeleton components, GTPases of the Rho family, and phospholipase C signaling pathway proteins. Phosphorylation of proteins involved in phagocytosis was only seen in response to TLR2 and TLR4 but not to TLR7 activation. Changes in the phosphorylation of proteins involved in endocytosis were delayed in response to TLR2 as compared to TLR4 ligands. These findings reveal that the phosphoproteomic response to stimulation of distinct TLRs varies both in the major modification targets and the phosphorylation dynamics. These results advance the understanding of how macrophages sense and respond to a diverse set of TLR stimuli.
Keywords: SILAC; TLR2; TLR4; TLR7; innate immunity; macrophage; phosphoproteomics; toll-like receptors.
Figures
Similar articles
-
A Methodology for Comprehensive Analysis of Toll-Like Receptor Signaling in Macrophages.Methods Mol Biol. 2017;1636:301-312. doi: 10.1007/978-1-4939-7154-1_19. Methods Mol Biol. 2017. PMID: 28730487 Free PMC article.
-
Phosphoproteome Analysis in Immune Cell Signaling.Curr Protoc Immunol. 2020 Sep;130(1):e105. doi: 10.1002/cpim.105. Curr Protoc Immunol. 2020. PMID: 32936995 Free PMC article.
-
Activation of Toll-like receptors by Burkholderia pseudomallei.BMC Immunol. 2008 Aug 8;9:46. doi: 10.1186/1471-2172-9-46. BMC Immunol. 2008. PMID: 18691413 Free PMC article.
-
Toll-like receptors as adjuvant receptors.Biochim Biophys Acta. 2002 Feb 13;1589(1):1-13. doi: 10.1016/s0167-4889(01)00182-3. Biochim Biophys Acta. 2002. PMID: 11909637 Review.
-
Charting protein dephosphorylation triggered by Toll-like receptor (TLR) signaling in macrophages and its role in health and disease.Int Rev Cell Mol Biol. 2021;361:211-243. doi: 10.1016/bs.ircmb.2021.02.003. Epub 2021 Mar 2. Int Rev Cell Mol Biol. 2021. PMID: 34074495 Review.
Cited by
-
Molecular mechanisms of macrophage Toll-like receptor-Fc receptor synergy.F1000Res. 2018 Jan 8;7:21. doi: 10.12688/f1000research.12679.1. eCollection 2018. F1000Res. 2018. PMID: 29375818 Free PMC article. Review.
-
Proteome and Secretome Analysis Reveals Differential Post-transcriptional Regulation of Toll-like Receptor Responses.Mol Cell Proteomics. 2017 Apr;16(4 suppl 1):S172-S186. doi: 10.1074/mcp.M116.064261. Epub 2017 Feb 24. Mol Cell Proteomics. 2017. PMID: 28235783 Free PMC article.
-
Differential regulation of the transcriptomic and secretomic landscape of sensor and effector functions of human airway epithelial cells.Mucosal Immunol. 2018 May;11(3):627-642. doi: 10.1038/mi.2017.100. Epub 2018 Jan 3. Mucosal Immunol. 2018. PMID: 29297499
-
Macrophage Phosphoproteome Analysis Reveals MINCLE-dependent and -independent Mycobacterial Cord Factor Signaling.Mol Cell Proteomics. 2019 Apr;18(4):669-685. doi: 10.1074/mcp.RA118.000929. Epub 2019 Jan 11. Mol Cell Proteomics. 2019. PMID: 30635358 Free PMC article.
-
Lipopolysaccharide Regulates the Macrophage RNA-Binding Proteome.J Proteome Res. 2024 Aug 2;23(8):3280-3293. doi: 10.1021/acs.jproteome.3c00838. Epub 2024 Mar 25. J Proteome Res. 2024. PMID: 38527097 Free PMC article.
References
-
- Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 2004, 4306996257–63. - PubMed
-
- Kang J. Y.; Lee J. O. Structural biology of the Toll-like receptor family. Annu. Rev. Biochem. 2011, 80, 917–41. - PubMed
-
- Werling D.; Jann O. C.; Offord V.; Glass E. J.; Coffey T. J. Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. 2009, 303124–30. - PubMed
-
- Takeda K.; Akira S. TLR signaling pathways. Semin. Immunol. 2004, 1613–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

