Analysis of human rotaviruses from a single location over an 18-year time span suggests that protein coadaption influences gene constellations
- PMID: 24942570
- PMCID: PMC4136370
- DOI: 10.1128/JVI.01562-14
Analysis of human rotaviruses from a single location over an 18-year time span suggests that protein coadaption influences gene constellations
Abstract
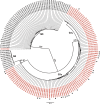
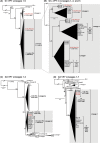
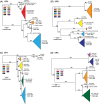
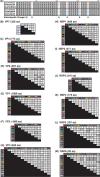
Rotaviruses (RVs) are 11-segmented, double-stranded RNA viruses that cause severe gastroenteritis in children. In addition to an error-prone genome replication mechanism, RVs can increase their genetic diversity by reassorting genes during host coinfection. Such exchanges allow RVs to acquire advantageous genes and adapt in the face of selective pressures. However, reassortment may also impose fitness costs if it unlinks genes/proteins that have accumulated compensatory, coadaptive mutations and that operate best when kept together. To better understand human RV evolutionary dynamics, we analyzed the genome sequences of 135 strains (genotype G1/G3/G4-P[8]-I1-C1-R1-A1-N1-T1-E1-H1) that were collected at a single location in Washington, DC, during the years 1974 to 1991. Intragenotypic phylogenetic trees were constructed for each viral gene using the nucleotide sequences, thereby defining novel allele level gene constellations (GCs) and illuminating putative reassortment events. The results showed that RVs with distinct GCs cocirculated during the vast majority of the collection years and that some of these GCs persisted in the community unchanged by reassortment. To investigate the influence of protein coadaptation on GC maintenance, we performed a mutual information-based analysis of the concatenated amino acid sequences and identified an extensive covariance network. Unexpectedly, amino acid covariation was highest between VP4 and VP2, which are structural components of the RV virion that are not thought to directly interact. These results suggest that GCs may be influenced by the selective constraints placed on functionally coadapted, albeit noninteracting, viral proteins. This work raises important questions about mutation-reassortment interplay and its impact on human RV evolution.
Importance: Rotaviruses are devastating human pathogens that cause severe diarrhea and kill >450,000 children each year. The virus can evolve by accumulating mutations and by acquiring new genes from other strains via a process called reassortment. However, little is known about the relationship between mutation accumulation and gene reassortment for rotaviruses and how it impacts viral evolution. In this study, we analyzed the genome sequences of human strains found in clinical fecal specimens that were collected at a single hospital over an 18-year time span. We found that many rotaviruses did not reassort their genes but instead maintained them as specific sets (i.e., constellations). By analyzing the encoded proteins, we discovered concurrent amino acid changes among them, which suggests that they are functionally coadapted to operate best when kept together. This study increases our understanding of how rotaviruses evolve over time in the human population.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures








Similar articles
-
Diversity and relationships of cocirculating modern human rotaviruses revealed using large-scale comparative genomics.J Virol. 2012 Sep;86(17):9148-62. doi: 10.1128/JVI.01105-12. Epub 2012 Jun 13. J Virol. 2012. PMID: 22696651 Free PMC article.
-
Intra-genotypic diversity of archival G4P[8] human rotaviruses from Washington, DC.Infect Genet Evol. 2011 Oct;11(7):1586-94. doi: 10.1016/j.meegid.2011.05.023. Epub 2011 Jun 25. Infect Genet Evol. 2011. PMID: 21712102 Free PMC article.
-
Molecular epidemiology of contemporary G2P[4] human rotaviruses cocirculating in a single U.S. community: footprints of a globally transitioning genotype.J Virol. 2014 Apr;88(7):3789-801. doi: 10.1128/JVI.03516-13. Epub 2014 Jan 15. J Virol. 2014. PMID: 24429371 Free PMC article.
-
Rotavirus diversity and evolution in the post-vaccine world.Discov Med. 2012 Jan;13(68):85-97. Discov Med. 2012. PMID: 22284787 Free PMC article. Review.
-
Equine Rotavirus A under the One Health Lens: Potential Impacts on Public Health.Viruses. 2024 Jan 16;16(1):130. doi: 10.3390/v16010130. Viruses. 2024. PMID: 38257830 Free PMC article. Review.
Cited by
-
Coding-Gene Coevolution Analysis of Rotavirus Proteins: A Bioinformatics and Statistical Approach.Genes (Basel). 2019 Dec 24;11(1):28. doi: 10.3390/genes11010028. Genes (Basel). 2019. PMID: 31878331 Free PMC article.
-
Genomic constellation of human Rotavirus A strains identified in Northern Brazil: a 6-year follow-up (2010-2016).Rev Inst Med Trop Sao Paulo. 2020 Dec 18;62:e98. doi: 10.1590/S1678-9946202062098. eCollection 2020. Rev Inst Med Trop Sao Paulo. 2020. PMID: 33331517 Free PMC article.
-
Reassortment in segmented RNA viruses: mechanisms and outcomes.Nat Rev Microbiol. 2016 Jul;14(7):448-60. doi: 10.1038/nrmicro.2016.46. Epub 2016 May 23. Nat Rev Microbiol. 2016. PMID: 27211789 Free PMC article. Review.
-
Distinguishing the genotype 1 genes and proteins of human Wa-like rotaviruses vs. porcine rotaviruses.Infect Genet Evol. 2016 Sep;43:6-14. doi: 10.1016/j.meegid.2016.05.014. Epub 2016 May 12. Infect Genet Evol. 2016. PMID: 27180895 Free PMC article.
-
Evolution of P[8], P[4], and P[6] VP8* genes of human rotaviruses globally reported during 1974 and 2017: possible implications for rotavirus vaccines in development.Hum Vaccin Immunother. 2019;15(12):3003-3008. doi: 10.1080/21645515.2019.1619400. Epub 2019 Jun 13. Hum Vaccin Immunother. 2019. PMID: 31124743 Free PMC article.
References
-
- Estes MK, Kapikian AZ. 2007. Rotaviruses and their replication, p 1917–1974 In Fields BN, Knipe DM, Howley PM, Griffen DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, PA
-
- Domingo E, Escarmis C, Sevilla N, Moya A, Elena SF, Quer J, Novella IS, Holland JJ. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859–864 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

