The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation
- PMID: 24942623
- PMCID: PMC4105754
- DOI: 10.1261/rna.046029.114
The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation
Abstract
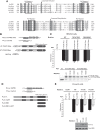
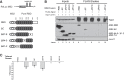
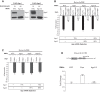
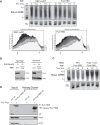
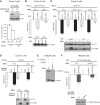
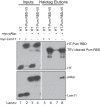
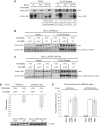
PUF proteins are potent repressors that serve important roles in stem cell maintenance, neurological processes, and embryonic development. These functions are driven by PUF protein recognition of specific binding sites within the 3' untranslated regions of target mRNAs. In this study, we investigated mechanisms of repression by the founding PUF, Drosophila Pumilio, and its human orthologs. Here, we evaluated a previously proposed model wherein the Pumilio RNA binding domain (RBD) binds Argonaute, which in turn blocks the translational activity of the eukaryotic elongation factor 1A. Surprisingly, we found that Argonautes are not necessary for repression elicited by Drosophila and human PUFs in vivo. A second model proposed that the RBD of Pumilio represses by recruiting deadenylases to shorten the mRNA's polyadenosine tail. Indeed, the RBD binds to the Pop2 deadenylase and accelerates deadenylation; however, this activity is not crucial for regulation. Rather, we determined that the poly(A) is necessary for repression by the RBD. Our results reveal that poly(A)-dependent repression by the RBD requires the poly(A) binding protein, pAbp. Furthermore, we show that repression by the human PUM2 RBD requires the pAbp ortholog, PABPC1. Pumilio associates with pAbp but does not disrupt binding of pAbp to the mRNA. Taken together, our data support a model wherein the Pumilio RBD antagonizes the ability of pAbp to promote translation. Thus, the conserved function of the PUF RBD is to bind specific mRNAs, antagonize pAbp function, and promote deadenylation.
Keywords: PUF; Pumilio; deadenylation; pAbp; poly(A) tail.
© 2014 Weidmann et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


Similar articles
-
Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs.J Biol Chem. 2012 Oct 19;287(43):36370-83. doi: 10.1074/jbc.M112.373522. Epub 2012 Sep 6. J Biol Chem. 2012. PMID: 22955276 Free PMC article.
-
Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes.EMBO J. 1996 Feb 15;15(4):900-9. EMBO J. 1996. PMID: 8631310 Free PMC article.
-
Levels of free PABP are limited by newly polyadenylated mRNA in early Spisula embryogenesis.Nucleic Acids Res. 2000 Sep 1;28(17):3346-53. doi: 10.1093/nar/28.17.3346. Nucleic Acids Res. 2000. PMID: 10954604 Free PMC article.
-
Roles of Puf proteins in mRNA degradation and translation.Wiley Interdiscip Rev RNA. 2011 Jul-Aug;2(4):471-92. doi: 10.1002/wrna.69. Epub 2010 Dec 16. Wiley Interdiscip Rev RNA. 2011. PMID: 21957038 Review.
-
LARP1 and LARP4: up close with PABP for mRNA 3' poly(A) protection and stabilization.RNA Biol. 2021 Feb;18(2):259-274. doi: 10.1080/15476286.2020.1868753. Epub 2021 Jan 31. RNA Biol. 2021. PMID: 33522422 Free PMC article. Review.
Cited by
-
Integrated analysis of RNA-binding protein complexes using in vitro selection and high-throughput sequencing and sequence specificity landscapes (SEQRS).Methods. 2017 Apr 15;118-119:171-181. doi: 10.1016/j.ymeth.2016.10.001. Epub 2016 Oct 8. Methods. 2017. PMID: 27729296 Free PMC article.
-
Pum2 Shapes the Transcriptome in Developing Axons through Retention of Target mRNAs in the Cell Body.Neuron. 2019 Dec 4;104(5):931-946.e5. doi: 10.1016/j.neuron.2019.08.035. Epub 2019 Oct 9. Neuron. 2019. PMID: 31606248 Free PMC article.
-
PTRE-seq reveals mechanism and interactions of RNA binding proteins and miRNAs.Nat Commun. 2018 Jan 19;9(1):301. doi: 10.1038/s41467-017-02745-0. Nat Commun. 2018. PMID: 29352242 Free PMC article.
-
RNA Degradation in Neurodegenerative Disease.Adv Neurobiol. 2018;20:103-142. doi: 10.1007/978-3-319-89689-2_5. Adv Neurobiol. 2018. PMID: 29916018 Free PMC article. Review.
-
Regulation of synapse density by Pumilio RNA-binding proteins.Cell Rep. 2024 Oct 22;43(10):114747. doi: 10.1016/j.celrep.2024.114747. Epub 2024 Sep 18. Cell Rep. 2024. PMID: 39298318 Free PMC article.
References
-
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S 1999. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol 1: 431–437 - PubMed
-
- Brook M, McCracken L, Reddington JP, Lu ZL, Morrice NA, Gray NK 2012. The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem J 441: 803–812 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
