Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection
- PMID: 24944192
- PMCID: PMC4326260
- DOI: 10.1126/scitranslmed.3007803
Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection
Abstract
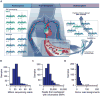
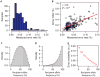
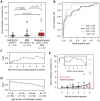
Monitoring allograft health is an important component of posttransplant therapy. Endomyocardial biopsy is the current gold standard for cardiac allograft monitoring but is an expensive and invasive procedure. Proof of principle of a universal, noninvasive diagnostic method based on high-throughput screening of circulating cell-free donor-derived DNA (cfdDNA) was recently demonstrated in a small retrospective cohort. We present the results of a prospective cohort study (65 patients, 565 samples) that tested the utility of cfdDNA in measuring acute rejection after heart transplantation. Circulating cell-free DNA was purified from plasma and sequenced (mean depth, 1.2 giga-base pairs) to quantify the fraction of cfdDNA. Through a comparison with endomyocardial biopsy results, we demonstrate that cfdDNA enables diagnosis of acute rejection after heart transplantation, with an area under the receiver operating characteristic curve of 0.83 and sensitivity and specificity that are comparable to the intrinsic performance of the biopsy itself. This noninvasive genome transplant dynamics approach is a powerful and informative method for routine monitoring of allograft health without incurring the risk, discomfort, and expense of an invasive biopsy.
Copyright © 2014, American Association for the Advancement of Science.
Conflict of interest statement
Figures

References
-
- Saraiva F, Matos V, Gonçalves L, Antunes M, Providência L. Complications of endomyocardial biopsy in heart transplant patients: A retrospective study of 2117 consecutive procedures. Transplant Proc. 2011;43:1908–1912. - PubMed
-
- Marboe CC, Billingham M, Eisen H, Deng MC, Baron H, Mehra M, Hunt S, Wohlgemuth J, Mahmood I, Prentice J, Berry G. Nodular endocardial infiltrates (Quilty lesions) cause significant variability in diagnosis of ISHLT grade 2 and 3A rejection in cardiac allograft recipients. J Heart Lung Transplant. 2005;24:S219–S226. - PubMed
-
- Zhang J, Tong KL, Li PK, Chan AY, Yeung CK, Pang CC, Wong TY, Lee KC, Lo YM. Presence of donor- and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: Urinary DNA chimerism. Clin Chem. 1999;45:1741–1746. - PubMed
-
- Lo YM, Tein MS, Pang CC, Yeung CK, Tong KL, Hjelm NM. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998;351:1329–1330. - PubMed
-
- Sigdel TK, Sarwal MM. Cell-free DNA as a measure of transplant injury. Clin Transpl. 2012:201–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

