The first endogenous herpesvirus, identified in the tarsier genome, and novel sequences from primate rhadinoviruses and lymphocryptoviruses
- PMID: 24945689
- PMCID: PMC4063692
- DOI: 10.1371/journal.pgen.1004332
The first endogenous herpesvirus, identified in the tarsier genome, and novel sequences from primate rhadinoviruses and lymphocryptoviruses
Abstract
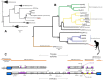
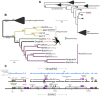
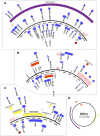
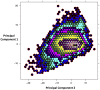
Herpesviridae is a diverse family of large and complex pathogens whose genomes are extremely difficult to sequence. This is particularly true for clinical samples, and if the virus, host, or both genomes are being sequenced for the first time. Although herpesviruses are known to occasionally integrate in host genomes, and can also be inherited in a Mendelian fashion, they are notably absent from the genomic fossil record comprised of endogenous viral elements (EVEs). Here, we combine paleovirological and metagenomic approaches to both explore the constituent viral diversity of mammalian genomes and search for endogenous herpesviruses. We describe the first endogenous herpesvirus from the genome of the Philippine tarsier, belonging to the Roseolovirus genus, and characterize its highly defective genome that is integrated and flanked by unambiguous host DNA. From a draft assembly of the aye-aye genome, we use bioinformatic tools to reveal over 100,000 bp of a novel rhadinovirus that is the first lemur gammaherpesvirus, closely related to Kaposi's sarcoma-associated virus. We also identify 58 genes of Pan paniscus lymphocryptovirus 1, the bonobo equivalent of human Epstein-Barr virus. For each of the viruses, we postulate gene function via comparative analysis to known viral relatives. Most notably, the evidence from gene content and phylogenetics suggests that the aye-aye sequences represent the most basal known rhadinovirus, and indicates that tumorigenic herpesviruses have been infecting primates since their emergence in the late Cretaceous. Overall, these data show that a genomic fossil record of herpesviruses exists despite their extremely large genomes, and expands the known diversity of Herpesviridae, which will aid the characterization of pathogenesis. Our analytical approach illustrates the benefit of intersecting evolutionary approaches with metagenomics, genetics and paleovirology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- McGeoch DJ, Davison AJ, Dolan A, Gatherer D, Sevilla-Reyes EE (2010) Molecular Evolution of the Herpesvirales. In: Domingo E, Holland JJ, editors. The Origin and Evolution of Viruses. Academic Press. 447–475 p. doi:10.1002/9780470688618.taw0208.
-
- Fields BN, Knipe DM, Howley PM (2007) Fields Virology. 5th ed. Philadelphia: Lippincott Williams and Wilkins.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

