Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microRNA dysregulation
- PMID: 24945770
- PMCID: PMC4086250
- DOI: 10.1016/j.neuron.2014.04.036
Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microRNA dysregulation
Abstract
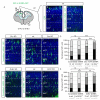
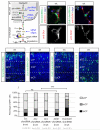
Acute gene inactivation using short hairpin RNA (shRNA, knockdown) in developing brain is a powerful technique to study genetic function; however, discrepancies between knockdown and knockout murine phenotypes have left unanswered questions. For example, doublecortin (Dcx) knockdown but not knockout shows a neocortical neuronal migration phenotype. Here we report that in utero electroporation of shRNA, but not siRNA or miRNA, to Dcx demonstrates a migration phenotype in Dcx knockouts akin to the effect in wild-type mice, suggesting shRNA-mediated off-target toxicity. This effect was not limited to Dcx, as it was observed in Dclk1 knockouts, as well as with a fraction of scrambled shRNAs, suggesting a sequence-dependent but not sequence-specific effect. Profiling RNAs from electroporated cells showed a defect in endogenous let7 miRNA levels, and disruption of let7 or Dicer recapitulated the migration defect. The results suggest that shRNA-mediated knockdown can produce untoward migration effects by altering endogenous miRNA pathways.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


References
-
- Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat Neurosci. 2003;6:1277–1283. - PubMed
-
- Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA biology. 2007;4:16–25. - PubMed
-
- Bauer M, Kinkl N, Meixner A, Kremmer E, Riemenschneider M, Forstl H, Gasser T, Ueffing M. Prevention of interferon-stimulated gene expression using microRNA-designed hairpins. Gene therapy. 2009;16:142–147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
