Quantification of HTLV-1 clonality and TCR diversity
- PMID: 24945836
- PMCID: PMC4063693
- DOI: 10.1371/journal.pcbi.1003646
Quantification of HTLV-1 clonality and TCR diversity
Abstract
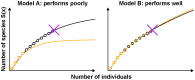
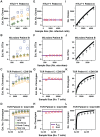
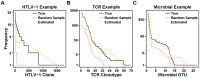
Estimation of immunological and microbiological diversity is vital to our understanding of infection and the immune response. For instance, what is the diversity of the T cell repertoire? These questions are partially addressed by high-throughput sequencing techniques that enable identification of immunological and microbiological "species" in a sample. Estimators of the number of unseen species are needed to estimate population diversity from sample diversity. Here we test five widely used non-parametric estimators, and develop and validate a novel method, DivE, to estimate species richness and distribution. We used three independent datasets: (i) viral populations from subjects infected with human T-lymphotropic virus type 1; (ii) T cell antigen receptor clonotype repertoires; and (iii) microbial data from infant faecal samples. When applied to datasets with rarefaction curves that did not plateau, existing estimators systematically increased with sample size. In contrast, DivE consistently and accurately estimated diversity for all datasets. We identify conditions that limit the application of DivE. We also show that DivE can be used to accurately estimate the underlying population frequency distribution. We have developed a novel method that is significantly more accurate than commonly used biodiversity estimators in microbiological and immunological populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Messaoudi I, Patino JAG, Dyall R, LeMaoult J, Nikolich-, et al (2002) Direct link between MHC polymorphism, T cell avidity, and diversity in immune defense. Science 298: 1797–1800. - PubMed
-
- Davenport MP, Price DA, McMichael AJ (2007) The T cell repertoire in infection and vaccination: implications for control of persistent viruses. Current Opinion in Immunology 19: 294–300. - PubMed
-
- Siegrist C-A, Aspinall R (2009) B-cell responses to vaccination at the extremes of age. Nat Rev Immunol 9: 185–194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

