Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation
- PMID: 24945926
- PMCID: PMC4063670
- DOI: 10.1371/journal.pgen.1004368
Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation
Abstract
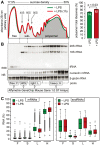
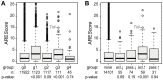
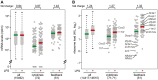
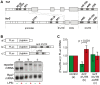
For a rapid induction and efficient resolution of the inflammatory response, gene expression in cells of the immune system is tightly regulated at the transcriptional and post-transcriptional level. The control of mRNA translation has emerged as an important determinant of protein levels, yet its role in macrophage activation is not well understood. We systematically analyzed the contribution of translational regulation to the early phase of the macrophage response by polysome fractionation from mouse macrophages stimulated with lipopolysaccharide (LPS). Individual mRNAs whose translation is specifically regulated during macrophage activation were identified by microarray analysis. Stimulation with LPS for 1 h caused translational activation of many feedback inhibitors of the inflammatory response including NF-κB inhibitors (Nfkbid, Nfkbiz, Nr4a1, Ier3), a p38 MAPK antagonist (Dusp1) and post-transcriptional suppressors of cytokine expression (Zfp36 and Zc3h12a). Our analysis showed that their translation is repressed in resting and de-repressed in activated macrophages. Quantification of mRNA levels at a high temporal resolution by RNASeq allowed us to define groups with different expression patterns. Thereby, we were able to distinguish mRNAs whose translation is actively regulated from mRNAs whose polysomal shifts are due to changes in mRNA levels. Active up-regulation of translation was associated with a higher content in AU-rich elements (AREs). For one example, Ier3 mRNA, we show that repression in resting cells as well as de-repression after stimulation depends on the ARE. Bone-marrow derived macrophages from Ier3 knockout mice showed reduced survival upon activation, indicating that IER3 induction protects macrophages from LPS-induced cell death. Taken together, our analysis reveals that translational control during macrophage activation is important for cellular survival as well as the expression of anti-inflammatory feedback inhibitors that promote the resolution of inflammation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Stamou P, Kontoyiannis DL (2010) Posttranscriptional regulation of TNF mRNA: a paradigm of signal-dependent mRNA utilization and its relevance to pathology. Curr Dir Autoimmun 11: 61–79. - PubMed
-
- Schott J, Stoecklin G (2010) Networks controlling mRNA decay in the immune system. Wiley Interdiscip Rev RNA 1: 432–456. - PubMed
-
- Garnon J, Lachance C, Di Marco S, Hel Z, Marion D, et al. (2005) Fragile X-related protein FXR1P regulates proinflammatory cytokine tumor necrosis factor expression at the post-transcriptional level. J Biol Chem 280: 5750–5763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

