Role of β-lactamase residues in a common interface for binding the structurally unrelated inhibitory proteins BLIP and BLIP-II
- PMID: 24947275
- PMCID: PMC4243995
- DOI: 10.1002/pro.2505
Role of β-lactamase residues in a common interface for binding the structurally unrelated inhibitory proteins BLIP and BLIP-II
Abstract
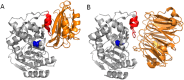
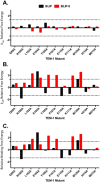
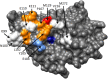
The β-lactamase inhibitory proteins (BLIPs) are a model system for examining molecular recognition in protein-protein interactions. BLIP and BLIP-II are structurally unrelated proteins that bind and inhibit TEM-1 β-lactamase. Both BLIPs share a common binding interface on TEM-1 and make contacts with many of the same TEM-1 surface residues. BLIP-II, however, binds TEM-1 over 150-fold tighter than BLIP despite the fact that it has fewer contact residues and a smaller binding interface. The role of eleven TEM-1 amino acid residues that contact both BLIP and BLIP-II was examined by alanine mutagenesis and determination of the association (k on) and dissociation (k off) rate constants for binding each partner. The substitutions had little impact on association rates and resulted in a wide range of dissociation rates as previously observed for substitutions on the BLIP side of the interface. The substitutions also had less effect on binding affinity for BLIP than BLIP-II. This is consistent with the high affinity and small binding interface of the TEM-1-BLIP-II complex, which predicts per residue contributions should be higher for TEM-1 binding to BLIP-II versus BLIP. Two TEM-1 residues (E104 and M129) were found to be hotspots for binding BLIP while five (L102, Y105, P107, K111, and M129) are hotspots for binding BLIP-II with only M129 as a common hotspot for both. Thus, although the same TEM-1 surface binds to both BLIP and BLIP-II, the distribution of binding energy on the surface is different for the two target proteins, that is, different binding strategies are employed.
Keywords: antibiotic resistance; beta-lactamase; binding kinetics; molecular recognition; protein-protein interactions.
© 2014 The Protein Society.
Figures


Similar articles
-
Engineering Specificity from Broad to Narrow: Design of a β-Lactamase Inhibitory Protein (BLIP) Variant That Exclusively Binds and Detects KPC β-Lactamase.ACS Infect Dis. 2016 Dec 9;2(12):969-979. doi: 10.1021/acsinfecdis.6b00160. Epub 2016 Oct 26. ACS Infect Dis. 2016. PMID: 27756125 Free PMC article.
-
Identification of the β-lactamase inhibitor protein-II (BLIP-II) interface residues essential for binding affinity and specificity for class A β-lactamases.J Biol Chem. 2013 Jun 14;288(24):17156-66. doi: 10.1074/jbc.M113.463521. Epub 2013 Apr 27. J Biol Chem. 2013. PMID: 23625930 Free PMC article.
-
BLIP-II Employs Differential Hotspot Residues To Bind Structurally Similar Staphylococcus aureus PBP2a and Class A β-Lactamases.Biochemistry. 2017 Feb 28;56(8):1075-1084. doi: 10.1021/acs.biochem.6b00978. Epub 2017 Feb 16. Biochemistry. 2017. PMID: 28182405 Free PMC article.
-
Insight into Structure-Function Relationships of β-Lactamase and BLIPs Interface Plasticity using Protein-Protein Interactions.Curr Pharm Des. 2019;25(31):3378-3389. doi: 10.2174/1381612825666190911154650. Curr Pharm Des. 2019. PMID: 31544712 Review.
-
Structure-based classification of class A beta-lactamases, an update.Curr Res Transl Med. 2019 Nov;67(4):115-122. doi: 10.1016/j.retram.2019.05.003. Epub 2019 May 31. Curr Res Transl Med. 2019. PMID: 31155436 Review.
Cited by
-
Tackling the Antibiotic Resistance Caused by Class A β-Lactamases through the Use of β-Lactamase Inhibitory Protein.Int J Mol Sci. 2018 Jul 30;19(8):2222. doi: 10.3390/ijms19082222. Int J Mol Sci. 2018. PMID: 30061509 Free PMC article.
-
Structural basis for the hydrolytic dehalogenation of the fungicide chlorothalonil.J Biol Chem. 2020 Jun 26;295(26):8668-8677. doi: 10.1074/jbc.RA120.013150. Epub 2020 Apr 30. J Biol Chem. 2020. PMID: 32358058 Free PMC article.
-
Tuning Electrostatic Gating of Semiconducting Carbon Nanotubes by Controlling Protein Orientation in Biosensing Devices.Angew Chem Weinheim Bergstr Ger. 2021 Sep 6;133(37):20346-20351. doi: 10.1002/ange.202104044. Epub 2021 Aug 6. Angew Chem Weinheim Bergstr Ger. 2021. PMID: 38504924 Free PMC article.
-
In vivo functional phenotypes from a computational epistatic model of evolution.Proc Natl Acad Sci U S A. 2024 Feb 6;121(6):e2308895121. doi: 10.1073/pnas.2308895121. Epub 2024 Jan 29. Proc Natl Acad Sci U S A. 2024. PMID: 38285950 Free PMC article.
-
Tuning Electrostatic Gating of Semiconducting Carbon Nanotubes by Controlling Protein Orientation in Biosensing Devices.Angew Chem Int Ed Engl. 2021 Sep 6;60(37):20184-20189. doi: 10.1002/anie.202104044. Epub 2021 Aug 6. Angew Chem Int Ed Engl. 2021. PMID: 34270157 Free PMC article.
References
-
- Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18:1233–1243. - PubMed
-
- Khan SH, Ahmad F, Ahmad N, Flynn DC, Kumar R. Protein-protein interactions: principles, techniques, and their potential role in new drug development. J Biomol Struct Dyn. 2011;28:929–938. - PubMed
-
- Zhang Z, Palzkill T. Determinants of binding affinity and specificity for the interaction of TEM-1 and SME-1 β-lactamase with β-lactamase inhibitory protein. J Biol Chem. 2003;278:45706–45712. - PubMed
-
- Zhang Z, Palzkill T. Dissecting the protein-protein interface between beta-lactamase inhibitory protein and class A beta-lactamases. J Biol Chem. 2004;279:42860–42866. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

