Nitrogen stress response and stringent response are coupled in Escherichia coli
- PMID: 24947454
- PMCID: PMC4066584
- DOI: 10.1038/ncomms5115
Nitrogen stress response and stringent response are coupled in Escherichia coli
Abstract
Assimilation of nitrogen is an essential process in bacteria. The nitrogen regulation stress response is an adaptive mechanism used by nitrogen-starved Escherichia coli to scavenge for alternative nitrogen sources and requires the global transcriptional regulator NtrC. In addition, nitrogen-starved E. coli cells synthesize a signal molecule, guanosine tetraphosphate (ppGpp), which serves as an effector molecule of many processes including transcription to initiate global physiological changes, collectively termed the stringent response. The regulatory mechanisms leading to elevated ppGpp levels during nutritional stresses remain elusive. Here, we show that transcription of relA, a key gene responsible for the synthesis of ppGpp, is activated by NtrC during nitrogen starvation. The results reveal that NtrC couples these two major bacterial stress responses to manage conditions of nitrogen limitation, and provide novel mechanistic insights into how a specific nutritional stress leads to elevating ppGpp levels in bacteria.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
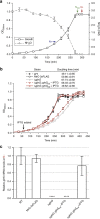
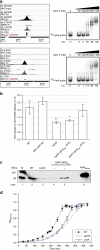

Comment in
-
Combinatorial stress responses: direct coupling of two major stress responses in Escherichia coli.Microb Cell. 2014 Sep 1;1(9):315-317. doi: 10.15698/mic2014.09.168. Microb Cell. 2014. PMID: 28357257 Free PMC article.
References
-
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57, 155–176 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

