The diabetes susceptibility gene Clec16a regulates mitophagy
- PMID: 24949970
- PMCID: PMC4184276
- DOI: 10.1016/j.cell.2014.05.016
The diabetes susceptibility gene Clec16a regulates mitophagy
Abstract
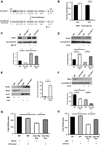
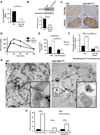
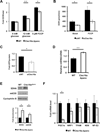
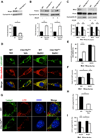
Clec16a has been identified as a disease susceptibility gene for type 1 diabetes, multiple sclerosis, and adrenal dysfunction, but its function is unknown. Here we report that Clec16a is a membrane-associated endosomal protein that interacts with E3 ubiquitin ligase Nrdp1. Loss of Clec16a leads to an increase in the Nrdp1 target Parkin, a master regulator of mitophagy. Islets from mice with pancreas-specific deletion of Clec16a have abnormal mitochondria with reduced oxygen consumption and ATP concentration, both of which are required for normal β cell function. Indeed, pancreatic Clec16a is required for normal glucose-stimulated insulin release. Moreover, patients harboring a diabetogenic SNP in the Clec16a gene have reduced islet Clec16a expression and reduced insulin secretion. Thus, Clec16a controls β cell function and prevents diabetes by controlling mitophagy. This pathway could be targeted for prevention and control of diabetes and may extend to the pathogenesis of other Clec16a- and Parkin-associated diseases.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

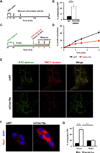
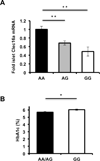
References
-
- Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J Biol Chem. 2006;281:38061–38070. - PubMed
-
- Chase HP, Cuthbertson DD, Dolan LM, Kaufman F, Krischer JP, Schatz DA, White NH, Wilson DM, Wolfsdorf J. First-phase insulin release during the intravenous glucose tolerance test as a risk factor for type 1 diabetes. J Pediatr. 2001;138:244–249. - PubMed
-
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

