Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles
- PMID: 24949977
- PMCID: PMC4087167
- DOI: 10.1016/j.cell.2014.04.038
Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles
Abstract
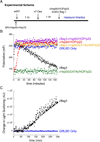
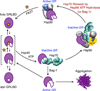
The glucocorticoid receptor (GR), like many signaling proteins, depends on the Hsp90 molecular chaperone for in vivo function. Although Hsp90 is required for ligand binding in vivo, purified apo GR is capable of binding ligand with no enhancement from Hsp90. We reveal that Hsp70, known to facilitate client delivery to Hsp90, inactivates GR through partial unfolding, whereas Hsp90 reverses this inactivation. Full recovery of ligand binding requires ATP hydrolysis on Hsp90 and the Hop and p23 cochaperones. Surprisingly, Hsp90 ATP hydrolysis appears to regulate client transfer from Hsp70, likely through a coupling of the two chaperone's ATP cycles. Such coupling is embodied in contacts between Hsp90 and Hsp70 in the GR:Hsp70:Hsp90:Hop complex imaged by cryoelectron microscopy. Whereas GR released from Hsp70 is aggregation prone, release from Hsp90 protects GR from aggregation and enhances its ligand affinity. Together, this illustrates how coordinated chaperone interactions can enhance stability, function, and regulation.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures





References
-
- Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear Receptor Structure: Implications for Function. Annu. Rev. Physiol. 2007;69:201–220. - PubMed
-
- Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM. Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel Mode of Receptor Dimerization and Coactivator Recognition. Cell. 2002;110:93–105. - PubMed
-
- Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 1998;273:35194–35200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

