Gene expression identifies heterogeneity of metastatic behavior among high-grade non-translocation associated soft tissue sarcomas
- PMID: 24950699
- PMCID: PMC4082412
- DOI: 10.1186/1479-5876-12-176
Gene expression identifies heterogeneity of metastatic behavior among high-grade non-translocation associated soft tissue sarcomas
Abstract
Background: The biologic heterogeneity of soft tissue sarcomas (STS), even within histological subtypes, complicates treatment. In earlier studies, gene expression patterns that distinguish two subsets of clear cell renal carcinoma (RCC), serous ovarian carcinoma (OVCA), and aggressive fibromatosis (AF) were used to separate 73 STS into two or four groups with different probabilities of developing metastatic disease (PrMet). This study was designed to confirm our earlier observations in a larger independent data set.
Methods: We utilized these gene sets, hierarchical clustering (HC), and Kaplan-Meier analysis, to examine 309 STS, using Affymetrix chip expression profiling.
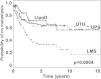
Results: HC using the combined AF-, RCC-, and OVCA-gene sets identified subsets of the STS samples. Analysis revealed differences in PrMet between the clusters defined by the first branch point of the clustering dendrogram (p = 0.048), and also among the four different clusters defined by the second branch points (p < 0.0001). Analysis also revealed differences in PrMet between the leiomyosarcomas (LMS), dedifferentiated liposarcomas (LipoD), and undifferentiated pleomorphic sarcomas (UPS) (p = 0.0004). HC of both the LipoD and UPS sample sets divided the samples into two groups with different PrMet (p = 0.0128, and 0.0002, respectively). HC of the UPS samples also showed four groups with different PrMet (p = 0.0007). HC found no subgroups of the LMS samples.
Conclusions: These data confirm our earlier studies, and suggest that this approach may allow the identification of more than two subsets of STS, each with distinct clinical behavior, and may be useful to stratify STS in clinical trials and in patient management.
Figures



References
-
- Helias-Rodzewicz Z, Pedeutour F, Coindre JM, Terrier P, Aurias A. Selective elimination of amplified CDK4 sequences correlates with spontaneous adipocytic differentiation in liposarcoma. Genes Chromosomes Cancer. 2009;48(11):943–952. - PubMed
-
- Ladanyi M, Bridge JA. Contribution of molecular genetic data to the classification of sarcomas. Hum Pathol. 2000;31(5):532–538. - PubMed
-
- Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462(2–3):247–253. - PubMed
-
- Skubitz KM, D’Adamo DR. Sarcoma. Mayo Clin Proc. 2007;82(11):1409–1432. - PubMed
-
- van de Rijn M, Fletcher JA. Genetics of soft tissue tumors. Annu Rev Pathol. 2006;1:435–466. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

