Evolution of the vertebrate Pax4/6 class of genes with focus on its novel member, the Pax10 gene
- PMID: 24951566
- PMCID: PMC4122933
- DOI: 10.1093/gbe/evu135
Evolution of the vertebrate Pax4/6 class of genes with focus on its novel member, the Pax10 gene
Abstract
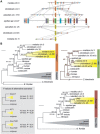
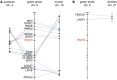
The members of the paired box (Pax) family regulate key developmental pathways in many metazoans as tissue-specific transcription factors. Vertebrate genomes typically possess nine Pax genes (Pax1-9), which are derived from four proto-Pax genes in the vertebrate ancestor that were later expanded through the so-called two-round (2R) whole-genome duplication. A recent study proposed that pax6a genes of a subset of teleost fishes (namely, acanthopterygians) are remnants of a paralog generated in the 2R genome duplication, to be renamed pax6.3, and reported one more group of vertebrate Pax genes (Pax6.2), most closely related to the Pax4/6 class. We propose to designate this new member Pax10 instead and reconstruct the evolutionary history of the Pax4/6/10 class with solid phylogenetic evidence. Our synteny analysis showed that Pax4, -6, and -10 originated in the 2R genome duplications early in vertebrate evolution. The phylogenetic analyses of relationships between teleost pax6a and other Pax4, -6, and -10 genes, however, do not support the proposed hypothesis of an ancient origin of the acanthopterygian pax6a genes in the 2R genome duplication. Instead, we confirmed the traditional scenario that the acanthopterygian pax6a is derived from the more recent teleost-specific genome duplication. Notably, Pax6 is present in all vertebrates surveyed to date, whereas Pax4 and -10 were lost multiple times in independent vertebrate lineages, likely because of their restricted expression patterns: Among Pax6-positive domains, Pax10 has retained expression in the adult retina alone, which we documented through in situ hybridization and quantitative reverse transcription polymerase chain reaction experiments on zebrafish, Xenopus, and anole lizard.
Keywords: Pax10; Pax4; Pax6; conserved synteny; gene duplication; gene loss.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures





Similar articles
-
Co-orthology of Pax4 and Pax6 to the fly eyeless gene: molecular phylogenetic, comparative genomic, and embryological analyses.Evol Dev. 2011 Sep-Oct;13(5):448-59. doi: 10.1111/j.1525-142X.2011.00502.x. Evol Dev. 2011. PMID: 23016906
-
Sequencing of Pax6 loci from the elephant shark reveals a family of Pax6 genes in vertebrate genomes, forged by ancient duplications and divergences.PLoS Genet. 2013;9(1):e1003177. doi: 10.1371/journal.pgen.1003177. Epub 2013 Jan 24. PLoS Genet. 2013. PMID: 23359656 Free PMC article.
-
Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence.PLoS Genet. 2008 Feb;4(2):e29. doi: 10.1371/journal.pgen.0040029. PLoS Genet. 2008. PMID: 18282108 Free PMC article.
-
Pax genes in eye development and evolution.Curr Opin Genet Dev. 2005 Aug;15(4):430-8. doi: 10.1016/j.gde.2005.05.001. Curr Opin Genet Dev. 2005. PMID: 15950457 Review.
-
Focus on molecules: Pax-6, the eye master.Exp Eye Res. 2006 Aug;83(2):233-4. doi: 10.1016/j.exer.2005.11.019. Epub 2006 Mar 23. Exp Eye Res. 2006. PMID: 16563385 Review. No abstract available.
Cited by
-
Lampreys, the jawless vertebrates, contain three Pax6 genes with distinct expression in eye, brain and pancreas.Sci Rep. 2019 Dec 20;9(1):19559. doi: 10.1038/s41598-019-56085-8. Sci Rep. 2019. PMID: 31863055 Free PMC article.
-
Homeodomain proteins: an update.Chromosoma. 2016 Jun;125(3):497-521. doi: 10.1007/s00412-015-0543-8. Epub 2015 Oct 13. Chromosoma. 2016. PMID: 26464018 Free PMC article. Review.
-
Evidence from cyclostomes for complex regionalization of the ancestral vertebrate brain.Nature. 2016 Mar 3;531(7592):97-100. doi: 10.1038/nature16518. Epub 2016 Feb 15. Nature. 2016. PMID: 26878236
-
Hagfish to Illuminate the Developmental and Evolutionary Origins of the Vertebrate Retina.Front Cell Dev Biol. 2022 Jan 26;10:822358. doi: 10.3389/fcell.2022.822358. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35155434 Free PMC article. Review.
-
Independent pseudogenizations and losses of sox15 during amniote diversification following asymmetric ohnolog evolution.BMC Ecol Evol. 2021 Jun 30;21(1):134. doi: 10.1186/s12862-021-01864-z. BMC Ecol Evol. 2021. PMID: 34193037 Free PMC article.
References
-
- Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. - PubMed
-
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

