LacR is a repressor of lacABCD and LacT is an activator of lacTFEG, constituting the lac gene cluster in Streptococcus pneumoniae
- PMID: 24951784
- PMCID: PMC4136080
- DOI: 10.1128/AEM.01370-14
LacR is a repressor of lacABCD and LacT is an activator of lacTFEG, constituting the lac gene cluster in Streptococcus pneumoniae
Abstract
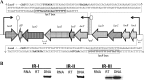
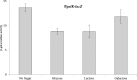
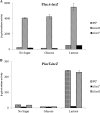
Comparison of the transcriptome of Streptococcus pneumoniae strain D39 grown in the presence of either lactose or galactose with that of the strain grown in the presence of glucose revealed the elevated expression of various genes and operons, including the lac gene cluster, which is organized into two operons, i.e., lac operon I (lacABCD) and lac operon II (lacTFEG). Deletion of the DeoR family transcriptional regulator lacR that is present downstream of the lac gene cluster revealed elevated expression of lac operon I even in the absence of lactose. This suggests a function of LacR as a transcriptional repressor of lac operon I, which encodes enzymes involved in the phosphorylated tagatose pathway in the absence of lactose or galactose. Deletion of lacR did not affect the expression of lac operon II, which encodes a lactose-specific phosphotransferase. This finding was further confirmed by β-galactosidase assays with PlacA-lacZ and PlacT-lacZ in the presence of either lactose or glucose as the sole carbon source in the medium. This suggests the involvement of another transcriptional regulator in the regulation of lac operon II, which is the BglG-family transcriptional antiterminator LacT. We demonstrate the role of LacT as a transcriptional activator of lac operon II in the presence of lactose and CcpA-independent regulation of the lac gene cluster in S. pneumoniae.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

