Application of coamplification at lower denaturation temperature-PCR sequencing for early detection of antiviral drug resistance mutations of hepatitis B virus
- PMID: 24951803
- PMCID: PMC4313139
- DOI: 10.1128/JCM.00343-14
Application of coamplification at lower denaturation temperature-PCR sequencing for early detection of antiviral drug resistance mutations of hepatitis B virus
Abstract
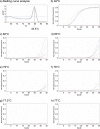
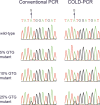
Nucleoside/nucleotide analogue for the treatment of chronic hepatitis B virus (HBV) infection is hampered by the emergence of drug resistance mutations. Conventional PCR sequencing cannot detect minor variants of <20%. We developed a modified co-amplification at lower denaturation temperature-PCR (COLD-PCR) method for the detection of HBV minority drug resistance mutations. The critical denaturation temperature for COLD-PCR was determined to be 78°C. Sensitivity of COLD-PCR sequencing was determined using serially diluted plasmids containing mixed proportions of HBV reverse transcriptase (rt) wild-type and mutant sequences. Conventional PCR sequencing detected mutations only if they existed in ≥25%, whereas COLD-PCR sequencing detected mutations when they existed in 5 to 10% of the viral population. The performance of COLD-PCR was compared to conventional PCR sequencing and a line probe assay (LiPA) using 215 samples obtained from 136 lamivudine- or telbivudine-treated patients with virological breakthrough. Among these 215 samples, drug resistance mutations were detected in 155 (72%), 148 (69%), and 113 samples (53%) by LiPA, COLD-PCR, and conventional PCR sequencing, respectively. Nineteen (9%) samples had mutations detectable by COLD-PCR but not LiPA, while 26 (12%) samples had mutations detectable by LiPA but not COLD-PCR, indicating both methods were comparable (P = 0.371). COLD-PCR was more sensitive than conventional PCR sequencing. Thirty-five (16%) samples had mutations detectable by COLD-PCR but not conventional PCR sequencing, while none had mutations detected by conventional PCR sequencing but not COLD-PCR (P < 0.0001). COLD-PCR sequencing is a simple method which is comparable to LiPA and superior to conventional PCR sequencing in detecting minor lamivudine/telbivudine resistance mutations.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Detection of hepatitis B virus genotypic resistance mutations by coamplification at lower denaturation temperature-PCR coupled with sanger sequencing.J Clin Microbiol. 2014 Aug;52(8):2933-9. doi: 10.1128/JCM.01127-14. Epub 2014 Jun 4. J Clin Microbiol. 2014. PMID: 24899029 Free PMC article.
-
Validation of the INNO-LiPA HBV DR assay (version 2) in monitoring hepatitis B virus-infected patients receiving nucleoside analog treatment.Antimicrob Agents Chemother. 2010 Mar;54(3):1283-9. doi: 10.1128/AAC.00970-09. Epub 2010 Jan 11. Antimicrob Agents Chemother. 2010. PMID: 20065049 Free PMC article.
-
Detection of rtN236T and rtA181V/T mutations associated with resistance to adefovir dipivoxil in samples from patients with chronic hepatitis B virus infection by the INNO-LiPA HBV DR line probe assay (version 2).J Clin Microbiol. 2006 Jun;44(6):1994-7. doi: 10.1128/JCM.02477-05. J Clin Microbiol. 2006. PMID: 16757589 Free PMC article.
-
Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay.J Clin Microbiol. 2002 Oct;40(10):3729-34. doi: 10.1128/JCM.40.10.3729-3734.2002. J Clin Microbiol. 2002. PMID: 12354872 Free PMC article.
-
PCR-based methods for the enrichment of minority alleles and mutations.Clin Chem. 2009 Apr;55(4):632-40. doi: 10.1373/clinchem.2008.113035. Epub 2009 Feb 6. Clin Chem. 2009. PMID: 19201784 Free PMC article. Review.
Cited by
-
COLD-PCR Method for Early Detection of Antiviral Drug-Resistance Mutations in Treatment-Naive Children with Chronic Hepatitis B.Diagnostics (Basel). 2020 Jul 18;10(7):491. doi: 10.3390/diagnostics10070491. Diagnostics (Basel). 2020. PMID: 32708399 Free PMC article.
-
COLD-PCR Technologies in the Area of Personalized Medicine: Methodology and Applications.Mol Diagn Ther. 2017 Jun;21(3):269-283. doi: 10.1007/s40291-016-0254-8. Mol Diagn Ther. 2017. PMID: 28101802 Review.
-
Eurotium cristatum from Fu Brick Tea Promotes Adipose Thermogenesis by Boosting Colonic Akkermansia muciniphila in High-Fat-Fed Obese Mice.Foods. 2023 Oct 10;12(20):3716. doi: 10.3390/foods12203716. Foods. 2023. PMID: 37893609 Free PMC article.
-
Fast and Sensitive Real-Time PCR Detection of Major Antiviral-Drug Resistance Mutations in Chronic Hepatitis B Patients by Use of a Predesigned Panel of Locked-Nucleic-Acid TaqMan Probes.J Clin Microbiol. 2021 Sep 20;59(10):e0093621. doi: 10.1128/JCM.00936-21. Epub 2021 Jul 28. J Clin Microbiol. 2021. PMID: 34319801 Free PMC article.
-
Detection of Anti-Hepatitis B Virus Drug Resistance Mutations Based on Multicolor Melting Curve Analysis.J Clin Microbiol. 2016 Nov;54(11):2661-2668. doi: 10.1128/JCM.00439-16. Epub 2016 Aug 17. J Clin Microbiol. 2016. PMID: 27535686 Free PMC article.
References
-
- Locarnini S, Hatzakis A, Heathcote J, Keeffe EB, Liang TJ, Mutimer D, Pawlotsky JM, Zoulim F. 2004. Management of antiviral resistance in patients with chronic hepatitis B. Antivir. Ther. 9:679–693. - PubMed
-
- Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, Liaw YF, Mizokami M, Kuiken C; Hepatitis B Virus Drug Resistance Working Group. 2007. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology 46:254–265. 10.1002/hep.21698. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

