A mass spectrometry view of stable and transient protein interactions
- PMID: 24952186
- PMCID: PMC5978421
- DOI: 10.1007/978-3-319-06068-2_11
A mass spectrometry view of stable and transient protein interactions
Abstract
Through an impressive range of dynamic interactions, proteins succeed to carry out the majority of functions in a cell. These temporally and spatially regulated interactions provide the means through which one single protein can perform diverse functions and modulate different cellular pathways. Understanding the identity and nature of these interactions is therefore critical for defining protein functions and their contribution to health and disease processes. Here, we provide an overview of workflows that incorporate immunoaffinity purifications and quantitative mass spectrometry (frequently abbreviated as IP-MS or AP-MS) for characterizing protein-protein interactions. We discuss experimental aspects that should be considered when optimizing the isolation of a protein complex. As the presence of nonspecific associations is a concern in these experiments, we discuss the common sources of nonspecific interactions and present label-free and metabolic labeling mass spectrometry-based methods that can help determine the specificity of interactions. The effective regulation of cellular pathways and the rapid reaction to various environmental stresses rely on the formation of stable, transient, and fast-exchanging protein-protein interactions. While determining the exact nature of an interaction remains challenging, we review cross-linking and metabolic labeling approaches that can help address this important aspect of characterizing protein interactions and macromolecular assemblies.
Figures
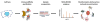

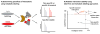
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

