Design of a colicin E7 based chimeric zinc-finger nuclease
- PMID: 24952471
- PMCID: PMC4104000
- DOI: 10.1007/s10822-014-9765-8
Design of a colicin E7 based chimeric zinc-finger nuclease
Abstract
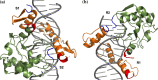
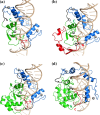
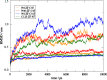
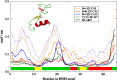
Colicin E7 is a natural bacterial toxin. Its nuclease domain (NColE7) enters the target cell and kills it by digesting the nucleic acids. The HNH-motif as the catalytic centre of NColE7 at the C-terminus requires the positively charged N-terminal loop for the nuclease activity-offering opportunities for allosteric control in a NColE7-based artificial nuclease. Accordingly, four novel zinc finger nucleases were designed by computational methods exploiting the special structural features of NColE7. The constructed models were subjected to MD simulations. The comparison of structural stability and functional aspects showed that these models may function as safely controlled artificial nucleases. This study was complemented by random mutagenesis experiments identifying potentially important residues for NColE7 function outside the catalytic region.
Figures




Similar articles
-
The role of the N-terminal loop in the function of the colicin E7 nuclease domain.J Biol Inorg Chem. 2013 Mar;18(3):309-21. doi: 10.1007/s00775-013-0975-7. Epub 2013 Jan 19. J Biol Inorg Chem. 2013. PMID: 23334162
-
Crystallization and preliminary crystallographic analysis of an Escherichia coli-selected mutant of the nuclease domain of the metallonuclease colicin E7.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 May 1;69(Pt 5):551-4. doi: 10.1107/S1744309113008233. Epub 2013 Apr 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23695575 Free PMC article.
-
The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis.Nucleic Acids Res. 2002 Apr 1;30(7):1670-8. doi: 10.1093/nar/30.7.1670. Nucleic Acids Res. 2002. PMID: 11917029 Free PMC article.
-
Importation of nuclease colicins into E coli cells: endoproteolytic cleavage and its prevention by the immunity protein.Biochimie. 2002 May-Jun;84(5-6):423-32. doi: 10.1016/s0300-9084(02)01426-8. Biochimie. 2002. PMID: 12423785 Review.
-
Nuclease colicins and their immunity proteins.Q Rev Biophys. 2012 Feb;45(1):57-103. doi: 10.1017/S0033583511000114. Epub 2011 Nov 16. Q Rev Biophys. 2012. PMID: 22085441 Review.
Cited by
-
Intrinsic protein disorder could be overlooked in cocrystallization conditions: An SRCD case study.Protein Sci. 2016 Nov;25(11):1977-1988. doi: 10.1002/pro.3010. Epub 2016 Aug 23. Protein Sci. 2016. PMID: 27508941 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

