Inferring human population size and separation history from multiple genome sequences
- PMID: 24952747
- PMCID: PMC4116295
- DOI: 10.1038/ng.3015
Inferring human population size and separation history from multiple genome sequences
Abstract
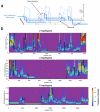
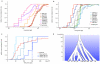
The availability of complete human genome sequences from populations across the world has given rise to new population genetic inference methods that explicitly model ancestral relationships under recombination and mutation. So far, application of these methods to evolutionary history more recent than 20,000-30,000 years ago and to population separations has been limited. Here we present a new method that overcomes these shortcomings. The multiple sequentially Markovian coalescent (MSMC) analyzes the observed pattern of mutations in multiple individuals, focusing on the first coalescence between any two individuals. Results from applying MSMC to genome sequences from nine populations across the world suggest that the genetic separation of non-African ancestors from African Yoruban ancestors started long before 50,000 years ago and give information about human population history as recent as 2,000 years ago, including the bottleneck in the peopling of the Americas and separations within Africa, East Asia and Europe.
Figures
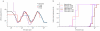
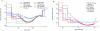
Comment in
-
Human evolution: multigenome coalescence for modern demographic insights.Nat Rev Genet. 2014 Aug;15(8):514. doi: 10.1038/nrg3781. Epub 2014 Jul 8. Nat Rev Genet. 2014. PMID: 25001845 No abstract available.
References
-
- Atkinson QD, Gray RD, Drummond AJ. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Molecular Biology and Evolution. 2008;25(2):468–474. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

