Genome sequences characterizing five mutations in RNA polymerase and major capsid of phages ϕA318 and ϕAs51 of Vibrio alginolyticus with different burst efficiencies
- PMID: 24952762
- PMCID: PMC4099482
- DOI: 10.1186/1471-2164-15-505
Genome sequences characterizing five mutations in RNA polymerase and major capsid of phages ϕA318 and ϕAs51 of Vibrio alginolyticus with different burst efficiencies
Abstract
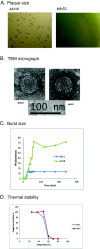
Background: The burst size of a phage is important prior to phage therapy and probiotic usage. The efficiency for a phage to burst its host bacterium can result from molecular domino effects of the phage gene expressions which dominate to control host machinery after infection. We found two Podoviridae phages, ϕA318 and ϕAs51, burst a common host V. alginolyticus with different efficiencies of 72 and 10 PFU/bacterium, respectively. Presumably, the genome sequences can be compared to explain their differences in burst sizes.
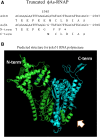
Results: Among genes in 42.5 kb genomes with a GC content of 43.5%, 16 out of 47 open-reading frames (ORFs) were annotated to known functions, including RNA polymerase (RNAP) and phage structure proteins. 11 strong phage promoters and three terminators were found. The consensus sequence for the new vibriophage promoters is AATAAAGTTGCCCTATA, where the AGTTG bases of -8 through -12 are important for the vibriophage specificity, especially a consensus T at -9 position eliminating RNAP of K1E, T7 and SP6 phages to transcribe the genes. ϕA318 and ϕAs51 RNAP shared their own specific promoters. In comparing ϕAs51 with ϕA318 genomes, only two nucleotides were deleted in the RNAP gene and three mutating nucleotides were found in the major capsid genes.
Conclusion: Subtle analyses on the residue alterations uncovered the effects of five nucleotide mutations on the functions of the RNAP and capsid proteins, which account for the host-bursting efficiency. The deletion of two nucleotides in RNAP gene truncates the primary translation due to early stop codon, while a second translational peptide starting from GTG just at deletion point can remediate the polymerase activity. Out of three nucleotide mutations in major capsid gene, H53N mutation weakens the subunit assembly between capsomeres for the phage head; E313K reduces the fold binding between β-sheet and Spine Helix inside the peptide.
Figures



References
-
- Baffone W, Citterio B, Vittoria E, Casaroli A, Pianetti A, Campana A, Bruscolini F. Determination of several potential virulence factors in Vibrio spp. isolated from seawater. Food Microbiol. 2001;18:479–488. doi: 10.1006/fmic.2001.0441. - DOI
-
- Kahla-Nakbi AB, Chaieb K, Besbes A, Zmantar T, Bakhrouf A. Virulence and enterobacterial repetitive intergenic consensus PCR of Vibrio alginolyticus strains isolated from Tunisian cultured gilthead sea bream and sea bass outbreaks. Vet Microbiol. 2006;117(2–4):321–327. doi: 10.1016/j.vetmic.2006.06.012. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

