A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain
- PMID: 24952961
- PMCID: PMC4319559
- DOI: 10.1016/j.neuron.2014.05.033
A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain
Abstract
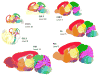
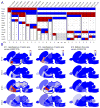
To provide a temporal framework for the genoarchitecture of brain development, we generated in situ hybridization data for embryonic and postnatal mouse brain at seven developmental stages for ∼2,100 genes, which were processed with an automated informatics pipeline and manually annotated. This resource comprises 434,946 images, seven reference atlases, an ontogenetic ontology, and tools to explore coexpression of genes across neurodevelopment. Gene sets coinciding with developmental phenomena were identified. A temporal shift in the principles governing the molecular organization of the brain was detected, with transient neuromeric, plate-based organization of the brain present at E11.5 and E13.5. Finally, these data provided a transcription factor code that discriminates brain structures and identifies the developmental age of a tissue, providing a foundation for eventual genetic manipulation or tracking of specific brain structures over development. The resource is available as the Allen Developing Mouse Brain Atlas (http://developingmouse.brain-map.org).
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures






References
-
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annual review of cell and developmental biology. 2009;25:45–69. - PubMed
-
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. - PubMed
-
- Bota M, Dong HW, Swanson LW. From gene networks to brain networks. Nat Neurosci. 2003;6:795–799. - PubMed
-
- Clancy B, Darlington RB, Finlay BL. Translating developmental time across mammalian species. Neuroscience. 2001;105:7–17. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

