Sex differences in the acute in vivo effects of different human SP-A variants on the mouse alveolar macrophage proteome
- PMID: 24954098
- PMCID: PMC4128237
- DOI: 10.1016/j.jprot.2014.06.007
Sex differences in the acute in vivo effects of different human SP-A variants on the mouse alveolar macrophage proteome
Abstract
Surfactant protein A (SP-A) is involved in lung innate immunity. Humans have two SP-A genes, SFTPA1 and SFTPA2, each with several variants. We examined the in vivo effects of treatment with specific SP-A variants on the alveolar macrophage (AM) proteome from SP-A knockout (KO) mice. KO mice received either SP-A1, SP-A2, or both. AM were collected and their proteomes examined with 2D-DIGE. We identified 90 proteins and categorized them as related to actin/cytoskeleton, oxidative stress, protease balance/chaperones, regulation of inflammation, and regulatory/developmental processes. SP-A1 and SP-A2 had different effects on the AM proteome and these effects differed between sexes. In males more changes occurred in the oxidative stress, protease/chaperones, and inflammation groups with SP-A2 treatment than with SP-A1. In females most SP-A1-induced changes were in the actin/cytoskeletal and oxidative stress groups. We conclude that after acute SP-A1 and SP-A2 treatment, sex-specific differences were observed in the AM proteomes from KO mice, and that these sex differences differ in response to SP-A1 and SP-A2. Females are more responsive to SP-A1, whereas the gene-specific differences in males were minimal. These observations not only demonstrate the therapeutic potential of exogenous SP-A, but also illustrate sex- and gene-specific differences in the response to it.
Biological significance: This study shows that changes occur in the alveolar macrophage proteome in response to a single in vivo treatment with exogenous SP-A1 and/or SP-A2. We demonstrate that SP-A1 and SP-A2 have different effects on the AM proteome and that sex differences exist in the response to each SP-A1 and SP-A2 gene product. This study illustrates the potential of exogenous SP-A1 and SP-A2 treatment for the manipulation of macrophage function and indicates that the specific SP-A variant used for treatment may vary with sex and with the cellular functions being modified. The observed changes may contribute to sex differences in the incidence of some lung diseases.
Keywords: Actin; Autophagy; Inflammation; Innate immunity; Oxidative stress; Surfactant.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures

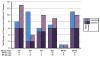








Similar articles
-
Sex-Specific Regulation of Gene Expression Networks by Surfactant Protein A (SP-A) Variants in Alveolar Macrophages in Response to Klebsiella pneumoniae.Front Immunol. 2020 Jun 24;11:1290. doi: 10.3389/fimmu.2020.01290. eCollection 2020. Front Immunol. 2020. PMID: 32670284 Free PMC article.
-
Survival of Surfactant Protein-A1 and SP-A2 Transgenic Mice After Klebsiella pneumoniae Infection, Exhibits Sex-, Gene-, and Variant Specific Differences; Treatment With Surfactant Protein Improves Survival.Front Immunol. 2018 Oct 16;9:2404. doi: 10.3389/fimmu.2018.02404. eCollection 2018. Front Immunol. 2018. PMID: 30459763 Free PMC article.
-
Differential Impact of Co-expressed SP-A1/SP-A2 Protein on AM miRNome; Sex Differences.Front Immunol. 2019 Aug 16;10:1960. doi: 10.3389/fimmu.2019.01960. eCollection 2019. Front Immunol. 2019. PMID: 31475015 Free PMC article.
-
Human Surfactant Protein SP-A1 and SP-A2 Variants Differentially Affect the Alveolar Microenvironment, Surfactant Structure, Regulation and Function of the Alveolar Macrophage, and Animal and Human Survival Under Various Conditions.Front Immunol. 2021 Aug 17;12:681639. doi: 10.3389/fimmu.2021.681639. eCollection 2021. Front Immunol. 2021. PMID: 34484180 Free PMC article. Review.
-
Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2--impact on function.Crit Rev Eukaryot Gene Expr. 2009;19(2):125-37. doi: 10.1615/critreveukargeneexpr.v19.i2.30. Crit Rev Eukaryot Gene Expr. 2009. PMID: 19392648 Free PMC article. Review.
Cited by
-
Can Prophylactic High Flow of Humidified and Warmed Filtered Air Improve Survival from Bacterial Pneumonia and SARS-CoV-2 in Elderly Individuals? The Role of Surfactant Protein A.Antioxidants (Basel). 2021 Apr 22;10(5):640. doi: 10.3390/antiox10050640. Antioxidants (Basel). 2021. PMID: 33922049 Free PMC article.
-
SP-A2 contributes to miRNA-mediated sex differences in response to oxidative stress: pro-inflammatory, anti-apoptotic, and anti-oxidant pathways are involved.Biol Sex Differ. 2017 Dec 4;8(1):37. doi: 10.1186/s13293-017-0158-2. Biol Sex Differ. 2017. PMID: 29202868 Free PMC article.
-
Sex and SP-A2 Dependent NAD(H) Redox Alterations in Mouse Alveolar Macrophages in Response to Ozone Exposure: Potential Implications for COVID-19.Antioxidants (Basel). 2020 Sep 25;9(10):915. doi: 10.3390/antiox9100915. Antioxidants (Basel). 2020. PMID: 32992843 Free PMC article.
-
The Lung Alveolar Cell (LAC) miRNome and Gene Expression Profile of the SP-A-KO Mice After Infection With and Without Rescue With Human Surfactant Protein-A2 (1A0).Front Immunol. 2022 Jul 1;13:854434. doi: 10.3389/fimmu.2022.854434. eCollection 2022. Front Immunol. 2022. PMID: 35844510 Free PMC article.
-
The Importance of Redox Status in the Frame of Lifestyle Approaches and the Genetics of the Lung Innate Immune Molecules, SP-A1 and SP-A2, on Differential Outcomes of COVID-19 Infection.Antioxidants (Basel). 2020 Aug 25;9(9):784. doi: 10.3390/antiox9090784. Antioxidants (Basel). 2020. PMID: 32854247 Free PMC article. Review.
References
-
- Madan T, Reid KB, Clark H, Singh M, Nayak A, Sarma PU, et al. Susceptibility of mice genetically deficient in SP-A or SP-D gene to invasive pulmonary aspergillosis. Mol Immunol. 2010;47:1923–1930. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

