Liver-specific transcriptional modules identified by genome-wide in silico analysis enable efficient gene therapy in mice and non-human primates
- PMID: 24954473
- PMCID: PMC4435486
- DOI: 10.1038/mt.2014.114
Liver-specific transcriptional modules identified by genome-wide in silico analysis enable efficient gene therapy in mice and non-human primates
Abstract
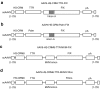
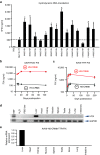
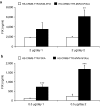
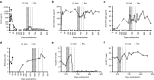
The robustness and safety of liver-directed gene therapy can be substantially improved by enhancing expression of the therapeutic transgene in the liver. To achieve this, we developed a new approach of rational in silico vector design. This approach relies on a genome-wide bio-informatics strategy to identify cis-acting regulatory modules (CRMs) containing evolutionary conserved clusters of transcription factor binding site motifs that determine high tissue-specific gene expression. Incorporation of these CRMs into adeno-associated viral (AAV) and non-viral vectors enhanced gene expression in mice liver 10 to 100-fold, depending on the promoter used. Furthermore, these CRMs resulted in robust and sustained liver-specific expression of coagulation factor IX (FIX), validating their immediate therapeutic and translational relevance. Subsequent translational studies indicated that therapeutic FIX expression levels could be attained reaching 20-35% of normal levels after AAV-based liver-directed gene therapy in cynomolgus macaques. This study underscores the potential of rational vector design using computational approaches to improve their robustness and therefore allows for the use of lower and thus safer vector doses for gene therapy, while maximizing therapeutic efficacy.
Figures


Comment in
-
Rational design for enhanced gene therapy with DNA transposons.Mol Ther. 2014 Sep;22(9):1575-7. doi: 10.1038/mt.2014.149. Mol Ther. 2014. PMID: 25186559 Free PMC article. No abstract available.
Similar articles
-
Genome-wide computational analysis reveals cardiomyocyte-specific transcriptional Cis-regulatory motifs that enable efficient cardiac gene therapy.Mol Ther. 2015 Jan;23(1):43-52. doi: 10.1038/mt.2014.178. Epub 2014 Sep 8. Mol Ther. 2015. PMID: 25195597 Free PMC article.
-
Computationally designed liver-specific transcriptional modules and hyperactive factor IX improve hepatic gene therapy.Blood. 2014 May 15;123(20):3195-9. doi: 10.1182/blood-2013-10-534032. Epub 2014 Mar 17. Blood. 2014. PMID: 24637359 Free PMC article.
-
Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose.Mol Ther. 2008 Feb;16(2):280-9. doi: 10.1038/sj.mt.6300355. Epub 2007 Dec 4. Mol Ther. 2008. PMID: 18059373
-
Theodore E. Woodward Award. AAV-mediated gene transfer for hemophilia.Trans Am Clin Climatol Assoc. 2003;114:337-51; discussion 351-2. Trans Am Clin Climatol Assoc. 2003. PMID: 12813929 Free PMC article. Review.
-
AAV-mediated gene transfer for hemophilia.Ann N Y Acad Sci. 2001 Dec;953:64-74. doi: 10.1111/j.1749-6632.2001.tb11361.x. Ann N Y Acad Sci. 2001. PMID: 11795424 Review.
Cited by
-
AAV vectors: The Rubik's cube of human gene therapy.Mol Ther. 2022 Dec 7;30(12):3515-3541. doi: 10.1016/j.ymthe.2022.09.015. Epub 2022 Oct 5. Mol Ther. 2022. PMID: 36203359 Free PMC article. Review.
-
Liver-targeted gene therapy: Approaches and challenges.Liver Transpl. 2015 Jun;21(6):718-37. doi: 10.1002/lt.24122. Liver Transpl. 2015. PMID: 25824605 Free PMC article. Review.
-
Moving forward toward a cure for hemophilia B.Mol Ther. 2015 May;23(5):809-811. doi: 10.1038/mt.2015.56. Mol Ther. 2015. PMID: 25943496 Free PMC article. No abstract available.
-
Update on clinical gene therapy for hemophilia.Blood. 2019 Jan 31;133(5):407-414. doi: 10.1182/blood-2018-07-820720. Epub 2018 Dec 17. Blood. 2019. PMID: 30559260 Free PMC article. Review.
-
AAV Vector-Mediated Antibody Delivery (A-MAD) in the Central Nervous System.Front Neurol. 2022 Apr 12;13:870799. doi: 10.3389/fneur.2022.870799. eCollection 2022. Front Neurol. 2022. PMID: 35493843 Free PMC article. Review.
References
-
- Kay MA. State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet. 2011;12:316–328. - PubMed
-
- Seymour LW, Thrasher AJ. Gene therapy matures in the clinic. Nat Biotechnol. 2012;30:588–593. - PubMed
-
- Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. - PubMed
-
- Mingozzi F, Maus MV, Hui DJ, Sabatino DE, Murphy SL, Rasko JE, et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat Med. 2007;13:419–422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

