Increasing the Coding Potential of Genomes Through Alternative Splicing: The Case of PARK2 Gene
- PMID: 24955028
- PMCID: PMC4064560
- DOI: 10.2174/1389202915666140426003342
Increasing the Coding Potential of Genomes Through Alternative Splicing: The Case of PARK2 Gene
Abstract
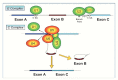
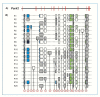
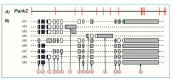
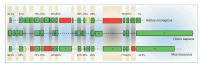
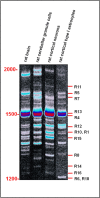
The completion of the Human Genome Project aroused renewed interest in alternative splicing, an efficient and widespread mechanism that generates multiple protein isoforms from individual genes. Although our knowledge about alternative splicing is growing exponentially, its real impact on cellular life is still to be clarified. Connecting all splicing features (genes, splice transcripts, isoforms, and relative functions) may be useful to resolve this tangle. Herein, we will start from the case of a single gene, Parkinson protein 2, E3 ubiquitin protein ligase (PARK2), one of the largest in our genome. This gene is implicated in the pathogenesis of autosomal recessive juvenile Parkinsonism and it has been recently linked to cancer, leprosy, autism, type 2 diabetes mellitus and Alzheimer's disease. PARK2 primary transcript undergoes an extensive alternative splicing, which enhances transcriptomic diversification and protein diversity in tissues and cells. This review will provide an update of all human PARK2 alternative splice transcripts and isoforms presently known, and correlate them to those in rat and mouse, two common animal models for studying human disease genes. Alternative splicing relies upon a complex process that could be easily altered by both cis and trans-acting mutations. Although the contribution of PARK2 splicing in human disease remains to be fully explored, some evidences show disruption of this versatile form of genetic regulation may have pathological consequences.
Keywords: Alternative splicing; PARK2; Protein isoforms; Splice expression patterns.; Splice variants; mRNA.
Figures

Similar articles
-
Alternative splicing generates different parkin protein isoforms: evidences in human, rat, and mouse brain.Biomed Res Int. 2014;2014:690796. doi: 10.1155/2014/690796. Epub 2014 Jul 16. Biomed Res Int. 2014. PMID: 25136611 Free PMC article.
-
Porcine Parkin: molecular cloning of PARK2 cDNA, expression analysis, and identification of a splicing variant.Biochem Biophys Res Commun. 2006 Sep 1;347(3):803-13. doi: 10.1016/j.bbrc.2006.06.167. Epub 2006 Jul 10. Biochem Biophys Res Commun. 2006. PMID: 16844087
-
Expanded alternative splice isoform profiling of the mouse Cav3.1/alpha1G T-type calcium channel.BMC Mol Biol. 2009 May 29;10:53. doi: 10.1186/1471-2199-10-53. BMC Mol Biol. 2009. PMID: 19480703 Free PMC article.
-
Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
-
Splicing: is there an alternative contribution to Parkinson's disease?Neurogenetics. 2015 Oct;16(4):245-63. doi: 10.1007/s10048-015-0449-x. Epub 2015 May 16. Neurogenetics. 2015. PMID: 25980689 Free PMC article. Review.
Cited by
-
Shifts in isoform usage underlie transcriptional differences in regulatory T cells in type 1 diabetes.Commun Biol. 2023 Sep 27;6(1):988. doi: 10.1038/s42003-023-05327-7. Commun Biol. 2023. PMID: 37758901 Free PMC article.
-
Copy number variability in Parkinson's disease: assembling the puzzle through a systems biology approach.Hum Genet. 2017 Jan;136(1):13-37. doi: 10.1007/s00439-016-1749-4. Epub 2016 Nov 28. Hum Genet. 2017. PMID: 27896429 Free PMC article. Review.
-
PARK2 inhibits osteosarcoma cell growth through the JAK2/STAT3/VEGF signaling pathway.Cell Death Dis. 2018 Mar 7;9(3):375. doi: 10.1038/s41419-018-0401-8. Cell Death Dis. 2018. PMID: 29515107 Free PMC article.
-
Aberrant transcriptional regulation could explain phenotypic variability in autosomal recessive polycystic kidney disease.J Mol Med (Berl). 2014 Oct;92(10):1011-4. doi: 10.1007/s00109-014-1197-3. J Mol Med (Berl). 2014. PMID: 25096485 No abstract available.
-
Proteomic Analysis of Parkin Isoforms Expression in Different Rat Brain Areas.Protein J. 2016 Oct;35(5):354-362. doi: 10.1007/s10930-016-9679-5. Protein J. 2016. PMID: 27601173
References
-
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. - PubMed
-
- Takahashi H, Ohama E, Suzuki S, Horikawa Y, Ishikawa A, Morita T, Tsuji S, Ikuta F. Familial juvenile parkinsonism: clinical and pathologic study in a family. Neurology. 1994;44(3 Pt 1):437–41. - PubMed
-
- Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V, D'Andrilli G, Scambia G, Picchio MC, Alder H, Godwin AK, Croce CM. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc. Natl. Acad. Sci. U S A. 2003;100(10):5956–61. - PMC - PubMed
-
- Veeriah S, Taylor B S, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan A J, Pao W, Ladanyi M, Sander C, Heguy A, Holland E C, Paty P B, Mischel P S, Liau L, Cloughesy T F, Melling-hoff I K, Solit D B, Chan T A. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat. Genet. 2010;42(1):77–82. - PMC - PubMed
-
- Mira M T, Alcais A, Nguyen V T, Moraes M O, Di Flumeri C, Vu H T, Mai C P, Nguyen T H, Nguyen N B, Pham X K, Sarno E N, Alter A, Montpetit A, Moraes M E, Moraes J R, Dore C, Gallant C J, Lepage P, Verner A, Van De Vosse E, Hudson T J, Abel L, Schurr E. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427(6975):636–40. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
