Ligand-binding pocket bridges DNA-binding and dimerization domains of the urate-responsive MarR homologue MftR from Burkholderia thailandensis
- PMID: 24955985
- PMCID: PMC4100783
- DOI: 10.1021/bi500219t
Ligand-binding pocket bridges DNA-binding and dimerization domains of the urate-responsive MarR homologue MftR from Burkholderia thailandensis
Abstract
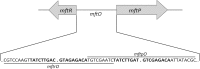
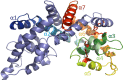
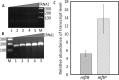
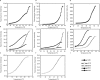
Members of the multiple antibiotic resistance regulator (MarR) family often regulate gene activity by responding to a specific ligand. In the absence of ligand, most MarR proteins function as repressors, while ligand binding causes attenuated DNA binding and therefore increased gene expression. Previously, we have shown that urate is a ligand for MftR (major facilitator transport regulator), which is encoded by the soil bacterium Burkholderia thailandensis. We show here that both mftR and the divergently oriented gene mftP encoding a major facilitator transport protein are upregulated in the presence of urate. MftR binds two cognate sites in the mftR-mftP intergenic region with equivalent affinity and sensitivity to urate. Mutagenesis of four conserved residues previously reported to be involved in urate binding to Deinococcus radiodurans HucR and Rhizobium radiobacter PecS significantly reduced protein stability and DNA binding affinity but not ligand binding. These data suggest that residues equivalent to those implicated in ligand binding to HucR and PecS serve structural roles and that MftR relies on distinct residues for ligand binding. MftR exhibits a two-step melting transition suggesting independent unfolding of the dimerization and DNA-binding regions; urate binding or mutations in the predicted ligand-binding sites result in one-step unfolding transitions. We suggest that MftR binds the ligand in a cleft between the DNA-binding lobes and the dimer interface but that the mechanism of ligand-mediated attenuation of DNA binding differs from that proposed for other urate-responsive MarR homologues. Since DNA binding by MftR is attenuated at 37 °C, our data also suggest that MftR responds to both ligand and a thermal upshift by attenuated DNA binding and upregulation of the genes under its control.
Figures


Similar articles
-
Urate-responsive MarR homologs from Burkholderia.Mol Biosyst. 2010 Nov;6(11):2133-42. doi: 10.1039/c0mb00086h. Epub 2010 Aug 23. Mol Biosyst. 2010. PMID: 20730247
-
Urate is a ligand for the transcriptional regulator PecS.J Mol Biol. 2010 Sep 24;402(3):539-51. doi: 10.1016/j.jmb.2010.07.053. Epub 2010 Aug 3. J Mol Biol. 2010. PMID: 20678501
-
MarR homologs with urate-binding signature.Protein Sci. 2011 Mar;20(3):621-9. doi: 10.1002/pro.588. Protein Sci. 2011. PMID: 21432936 Free PMC article.
-
MarR family transcription factors: dynamic variations on a common scaffold.Crit Rev Biochem Mol Biol. 2017 Dec;52(6):595-613. doi: 10.1080/10409238.2017.1344612. Epub 2017 Jul 3. Crit Rev Biochem Mol Biol. 2017. PMID: 28670937 Review.
-
MarR Family Transcription Factors from Burkholderia Species: Hidden Clues to Control of Virulence-Associated Genes.Microbiol Mol Biol Rev. 2018 Nov 28;83(1):e00039-18. doi: 10.1128/MMBR.00039-18. Print 2019 Mar. Microbiol Mol Biol Rev. 2018. PMID: 30487164 Free PMC article. Review.
Cited by
-
Secondary metabolites from the Burkholderia pseudomallei complex: structure, ecology, and evolution.J Ind Microbiol Biotechnol. 2020 Oct;47(9-10):877-887. doi: 10.1007/s10295-020-02317-0. Epub 2020 Oct 14. J Ind Microbiol Biotechnol. 2020. PMID: 33052546 Free PMC article. Review.
-
A MarR family transcriptional regulator and subinhibitory antibiotics regulate type VI secretion gene clusters in Burkholderia pseudomallei.Microbiology (Reading). 2018 Sep;164(9):1196-1211. doi: 10.1099/mic.0.000697. Epub 2018 Jul 27. Microbiology (Reading). 2018. PMID: 30052173 Free PMC article.
-
Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis.Sci Rep. 2017 Jul 25;7(1):6471. doi: 10.1038/s41598-017-01705-4. Sci Rep. 2017. PMID: 28743871 Free PMC article.
-
An EmrB multidrug efflux pump in Burkholderia thailandensis with unexpected roles in antibiotic resistance.J Biol Chem. 2019 Feb 8;294(6):1891-1903. doi: 10.1074/jbc.RA118.006638. Epub 2018 Dec 13. J Biol Chem. 2019. PMID: 30545940 Free PMC article.
-
Regulation of Metabolic Pathways by MarR Family Transcription Factors.Comput Struct Biotechnol J. 2017 Jun 16;15:366-371. doi: 10.1016/j.csbj.2017.06.001. eCollection 2017. Comput Struct Biotechnol J. 2017. PMID: 28694934 Free PMC article. Review.
References
-
- Wilkinson S. P.; Grove A. (2006) Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 8, 51–62. - PubMed
-
- Ellison D. W.; Miller V. L. (2006) Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9, 153–159. - PubMed
-
- Perera I. C.; Grove A. (2010) Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2, 243–254. - PubMed
-
- Grove A. (2013) MarR family transcription factors. Curr. Biol. 23, R142–143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

