Tissue-specific expressed antibody variable gene repertoires
- PMID: 24956460
- PMCID: PMC4067404
- DOI: 10.1371/journal.pone.0100839
Tissue-specific expressed antibody variable gene repertoires
Erratum in
-
Correction: Tissue-Specific Expressed Antibody Variable Gene Repertoires.PLoS One. 2020 Jan 24;15(1):e0228412. doi: 10.1371/journal.pone.0228412. eCollection 2020. PLoS One. 2020. PMID: 31978140 Free PMC article.
Abstract
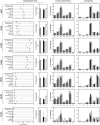
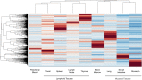
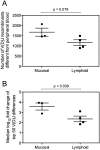
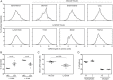
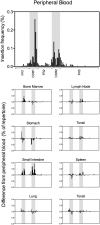
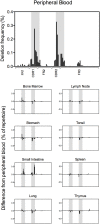
Recent developments in genetic technologies allow deep analysis of the sequence diversity of immune repertoires, but little work has been reported on the architecture of immune repertoires in mucosal tissues. Antibodies are the key to prevention of infections at the mucosal surface, but it is currently unclear whether the B cell repertoire at mucosal surfaces reflects the dominant antibodies found in the systemic compartment or whether mucosal tissues harbor unique repertoires. We examined the expressed antibody variable gene repertoires from 10 different human tissues using RNA samples derived from a large number of individuals. The results revealed that mucosal tissues such as stomach, intestine and lung possess unique antibody gene repertoires that differed substantially from those found in lymphoid tissues or peripheral blood. Mutation frequency analysis of mucosal tissue repertoires revealed that they were highly mutated, with little evidence for the presence of naïve B cells, in contrast to blood. Mucosal tissue repertoires possessed longer heavy chain complementarity determining region 3 loops than lymphoid tissue repertoires. We also noted a large increase in frequency of both insertions and deletions in the small intestine antibody repertoire. These data suggest that mucosal immune repertoires are distinct in many ways from the systemic compartment.
Conflict of interest statement
Figures

References
-
- Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302: 575–581. - PubMed
-
- Alt FW, Oltz EM, Young F, Gorman J, Taccioli G, et al. (1992) VDJ recombination. Immunology Today 13: 306–314. - PubMed
-
- Milstein C, Neuberger M (1996) Maturation of the immune response. Adv Protein Chem 49: 451–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

