IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family
- PMID: 24956979
- PMCID: PMC4141478
- DOI: 10.1038/ncomms5178
IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family
Abstract
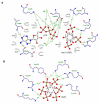
Inositol trisphosphate kinases (IP3Ks) and inositol hexakisphosphate kinases (IP6Ks) each regulate specialized signalling activities by phosphorylating either InsP3 or InsP6 respectively. The molecular basis for these different kinase activities can be illuminated by a structural description of IP6K. Here we describe the crystal structure of an Entamoeba histolytica hybrid IP6K/IP3K, an enzymatic parallel to a 'living fossil'. Through molecular modelling and mutagenesis, we extrapolated our findings to human IP6K2, which retains vestigial IP3K activity. Two structural elements, an α-helical pair and a rare, two-turn 310 helix, together forge a substrate-binding pocket with an open clamshell geometry. InsP6 forms substantial contacts with both structural elements. Relative to InsP6, enzyme-bound InsP3 rotates 55° closer to the α-helices, which provide most of the protein's interactions with InsP3. These data reveal the molecular determinants of IP6K activity, and suggest an unusual evolutionary trajectory for a primordial kinase that could have favored efficient bifunctionality, before propagation of separate IP3Ks and IP6Ks.
Figures




References
-
- Wilson MS, Livermore TM, Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 2013;452:369–379. - PubMed
-
- Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–805. - PubMed
-
- Illies C, et al. Inositol pyrophosphates determine exocytic capacity. Science. 2007;318:1299–1302. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

