Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor-positive breast cancer
- PMID: 24957076
- PMCID: PMC4110474
- DOI: 10.1093/jnci/dju158
Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor-positive breast cancer
Abstract
Background: Obesity increases the risk of cancer death among postmenopausal women with estrogen receptor-positive (ER+) breast cancer, but the direct evidence for the mechanisms is lacking. The purpose of this study is to demonstrate direct evidence for the mechanisms mediating this epidemiologic phenomenon.
Methods: We analyzed transcriptomic profiles of pretreatment biopsies from a prospective cohort of 137 ER+ breast cancer patients. We generated transgenic (MMTV-TGFα;A (y) /a) and orthotopic/syngeneic (A (y) /a) obese mouse models to investigate the effect of obesity on tumorigenesis and tumor progression and to determine biological mechanisms using whole-genome transcriptome microarrays and protein analyses. We used a coculture system to examine the impact of adipocytes/adipokines on breast cancer cell proliferation. All statistical tests were two-sided.
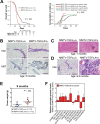
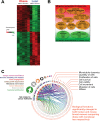
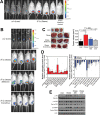
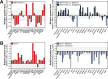
Results: Functional transcriptomic analysis of patients revealed the association of obesity with 59 biological functional changes (P < .05) linked to cancer hallmarks. Gene enrichment analysis revealed enrichment of AKT-target genes (P = .04) and epithelial-mesenchymal transition genes (P = .03) in patients. Our obese mouse models demonstrated activation of the AKT/mTOR pathway in obesity-accelerated mammary tumor growth (3.7- to 7.0-fold; P < .001; n = 6-7 mice per group). Metformin or everolimus can suppress obesity-induced secretion of adipokines and breast tumor formation and growth (0.5-fold, P = .04; 0.3-fold, P < .001, respectively; n = 6-8 mice per group). The coculture model revealed that adipocyte-secreted adipokines (eg, TIMP-1) regulate adipocyte-induced breast cancer cell proliferation and invasion. Metformin suppress adipocyte-induced cell proliferation and adipocyte-secreted adipokines in vitro.
Conclusions: Adipokine secretion and AKT/mTOR activation play important roles in obesity-accelerated breast cancer aggressiveness in addition to hyperinsulinemia, estrogen signaling, and inflammation. Metformin and everolimus have potential for therapeutic interventions of ER+ breast cancer patients with obesity.
© The Author 2014. Published by Oxford University Press.
Figures


References
-
- Renehan AG, Tyson M, Egger M, et al. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371(9612):569–578 - PubMed
-
- Petrelli JM, Calle EE, Rodriguez C, et al. Body mass index, height, and postmenopausal breast cancer mortality in a prospective cohort of US women. Cancer Causes Control. 2002;13(4):325–332 - PubMed
-
- Carmichael AR. Obesity as a risk factor for development and poor prognosis of breast cancer. BJOG. 2006;113(10):1160–1166 - PubMed
-
- Renehan AG, Soerjomataram I, Tyson M, et al. Incident cancer burden attributable to excess body mass index in 30 European countries. Int J Cancer. 2010;126(3):692–702 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

