Variable recombination dynamics during the emergence, transmission and 'disarming' of a multidrug-resistant pneumococcal clone
- PMID: 24957517
- PMCID: PMC4094930
- DOI: 10.1186/1741-7007-12-49
Variable recombination dynamics during the emergence, transmission and 'disarming' of a multidrug-resistant pneumococcal clone
Abstract
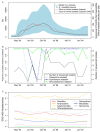
Background: Pneumococcal β-lactam resistance was first detected in Iceland in the late 1980s, and subsequently peaked at almost 25% of clinical isolates in the mid-1990s largely due to the spread of the internationally-disseminated multidrug-resistant PMEN2 (or Spain6B-2) clone of Streptococcus pneumoniae.
Results: Whole genome sequencing of an international collection of 189 isolates estimated that PMEN2 emerged around the late 1960s, developing resistance through multiple homologous recombinations and the acquisition of a Tn5253-type integrative and conjugative element (ICE). Two distinct clades entered Iceland in the 1980s, one of which had acquired a macrolide resistance cassette and was estimated to have risen sharply in its prevalence by coalescent analysis. Transmission within the island appeared to mainly emanate from Reykjavík and the Southern Peninsular, with evolution of the bacteria effectively clonal, mainly due to a prophage disrupting a gene necessary for genetic transformation in many isolates. A subsequent decline in PMEN2's prevalence in Iceland coincided with a nationwide campaign that reduced dispensing of antibiotics to children in an attempt to limit its spread. Specific mutations causing inactivation or loss of ICE-borne resistance genes were identified from the genome sequences of isolates that reverted to drug susceptible phenotypes around this time. Phylogenetic analysis revealed some of these occurred on multiple occasions in parallel, suggesting they may have been at least temporarily advantageous. However, alteration of 'core' sequences associated with resistance was precluded by the absence of any substantial homologous recombination events.
Conclusions: PMEN2's clonal evolution was successful over the short-term in a limited geographical region, but its inability to alter major antigens or 'core' gene sequences associated with resistance may have prevented persistence over longer timespans.
Figures




References
-
- Klugman KP. The successful clone: the vector of dissemination of resistance inStreptococcus pneumoniae. J Antimicrob Chemother. 2002;12:1–5. - PubMed
-
- Jacobs MR, Koornhof HJ, Robins-Browne RM, Stevenson CM, Vermaak ZA, Freiman I, Miller GB, Witcomb MA, Isaacson M, Ward JI, Austrian R. Emergence of multiply resistant pneumococci. N Engl J Med. 1978;12:735–740. - PubMed
-
- Fenoll A, Martin Bourgon C, Munoz R, Vicioso D, Casal J. Serotype distribution and antimicrobial resistance ofStreptococcus pneumoniaeisolates causing systemic infections in Spain, 1979–1989. Rev Infect Dis. 1991;12:56–60. - PubMed
-
- Marton A, Gulyas M, Munoz R, Tomasz A. Extremely high incidence of antibiotic resistance in clinical isolates of Streptococcus pneumoniae in Hungary. J Infect Dis. 1991;12:542–548. - PubMed
-
- Kristinsson KG, Hjalmarsdottir MA, Steingrimsson O. Increasing penicillin resistance in pneumococci in Iceland. Lancet. 1992;12:1606. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

