The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems
- PMID: 24957599
- PMCID: PMC4117751
- DOI: 10.1093/nar/gku500
The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems
Abstract
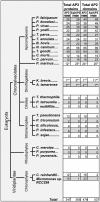
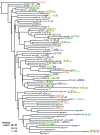
We provide the first comprehensive analysis of any transcription factor family in Cryptosporidium, a basal-branching apicomplexan that is the second leading cause of infant diarrhea globally. AP2 domain-containing proteins have evolved to be the major regulatory family in the phylum to the exclusion of canonical regulators. We show that apicomplexan and perkinsid AP2 domains cluster distinctly from other chromalveolate AP2s. Protein-binding specificity assays of C. parvum AP2 domains combined with motif conservation upstream of co-regulated gene clusters allowed the construction of putative AP2 regulons across the in vitro life cycle. Orthologous Apicomplexan AP2 (ApiAP2) expression has been rearranged relative to the malaria parasite P. falciparum, suggesting ApiAP2 network rewiring during evolution. C. hominis orthologs of putative C. parvum ApiAP2 proteins and target genes show greater than average variation. C. parvum AP2 domains display reduced binding diversity relative to P. falciparum, with multiple domains binding the 5'-TGCAT-3', 5'-CACACA-3' and G-box motifs (5'-G[T/C]GGGG-3'). Many overrepresented motifs in C. parvum upstream regions are not AP2 binding motifs. We propose that C. parvum is less reliant on ApiAP2 regulators in part because it utilizes E2F/DP1 transcription factors. C. parvum may provide clues to the ancestral state of apicomplexan transcriptional regulation, pre-AP2 domination.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains.Nucleic Acids Res. 2005 Jul 21;33(13):3994-4006. doi: 10.1093/nar/gki709. Print 2005. Nucleic Acids Res. 2005. PMID: 16040597 Free PMC article.
-
Genome-wide upstream motif analysis of Cryptosporidium parvum genes clustered by expression profile.BMC Genomics. 2013 Jul 29;14:516. doi: 10.1186/1471-2164-14-516. BMC Genomics. 2013. PMID: 23895416 Free PMC article.
-
Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite.PLoS Pathog. 2010 Oct 28;6(10):e1001165. doi: 10.1371/journal.ppat.1001165. PLoS Pathog. 2010. PMID: 21060817 Free PMC article.
-
Unraveling the complexities of ApiAP2 regulation in Plasmodium falciparum.Trends Parasitol. 2024 Nov;40(11):987-999. doi: 10.1016/j.pt.2024.09.007. Epub 2024 Oct 16. Trends Parasitol. 2024. PMID: 39419713 Review.
-
The Apicomplexan AP2 family: integral factors regulating Plasmodium development.Mol Biochem Parasitol. 2011 Mar;176(1):1-7. doi: 10.1016/j.molbiopara.2010.11.014. Epub 2010 Nov 30. Mol Biochem Parasitol. 2011. PMID: 21126543 Free PMC article. Review.
Cited by
-
The Modular Circuitry of Apicomplexan Cell Division Plasticity.Front Cell Infect Microbiol. 2021 Apr 12;11:670049. doi: 10.3389/fcimb.2021.670049. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33912479 Free PMC article. Review.
-
Sexual development in Plasmodium parasites: knowing when it's time to commit.Nat Rev Microbiol. 2015 Sep;13(9):573-87. doi: 10.1038/nrmicro3519. Nat Rev Microbiol. 2015. PMID: 26272409 Review.
-
Opposing Transcriptional Mechanisms Regulate Toxoplasma Development.mSphere. 2017 Feb 22;2(1):e00347-16. doi: 10.1128/mSphere.00347-16. eCollection 2017 Jan-Feb. mSphere. 2017. PMID: 28251183 Free PMC article.
-
Comparative genomics reveals Cyclospora cayetanensis possesses coccidia-like metabolism and invasion components but unique surface antigens.BMC Genomics. 2016 Apr 30;17:316. doi: 10.1186/s12864-016-2632-3. BMC Genomics. 2016. PMID: 27129308 Free PMC article.
-
Essential Genes of the Parasitic Apicomplexa.Trends Parasitol. 2021 Apr;37(4):304-316. doi: 10.1016/j.pt.2020.11.007. Epub 2021 Jan 5. Trends Parasitol. 2021. PMID: 33419671 Free PMC article. Review.
References
-
- Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., Wu Y., Sow S.O., Sur D., Breiman R.F., et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. - PubMed
-
- Okamoto N., McFadden G.I. The mother of all parasites. Future Microbiol. 2008;3:391–395.
-
- Putnam N.H., Butts T., Ferrier D.E., Furlong R.F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

