Systematic applications of metabolomics in metabolic engineering
- PMID: 24957776
- PMCID: PMC3901235
- DOI: 10.3390/metabo2041090
Systematic applications of metabolomics in metabolic engineering
Abstract
The goals of metabolic engineering are well-served by the biological information provided by metabolomics: information on how the cell is currently using its biochemical resources is perhaps one of the best ways to inform strategies to engineer a cell to produce a target compound. Using the analysis of extracellular or intracellular levels of the target compound (or a few closely related molecules) to drive metabolic engineering is quite common. However, there is surprisingly little systematic use of metabolomics datasets, which simultaneously measure hundreds of metabolites rather than just a few, for that same purpose. Here, we review the most common systematic approaches to integrating metabolite data with metabolic engineering, with emphasis on existing efforts to use whole-metabolome datasets. We then review some of the most common approaches for computational modeling of cell-wide metabolism, including constraint-based models, and discuss current computational approaches that explicitly use metabolomics data. We conclude with discussion of the broader potential of computational approaches that systematically use metabolomics data to drive metabolic engineering.
Figures
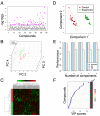
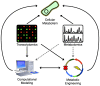
References
-
- Zaldivar J.Z., Borges A.B., Johansson B.J., Smits H.S., Villas-Bôas S.V.-B., Nielsen J.N., Olsson L.O. Fermentation performance and intracellular metabolite patterns in laboratory and industrial xylose-fermenting Saccharomyces cerevisiae. Appl. Microbiol. Biot. 2002;59:436–442. doi: 10.1007/s00253-002-1056-y. - DOI - PubMed
-
- Sonderegger M., Jeppsson M., Hahn-Hägerdal B., Sauer U. Molecular Basis for Anaerobic Growth of Saccharomyces cerevisiae on Xylose, Investigated by Global Gene Expression and Metabolic Flux Analysis. Appl. Environ. Microb. 2004;70:2307–2317. doi: 10.1128/AEM.70.4.2307-2317.2004. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

