SVA retrotransposon insertion-associated deletion represents a novel mutational mechanism underlying large genomic copy number changes with non-recurrent breakpoints
- PMID: 24958239
- PMCID: PMC4229983
- DOI: 10.1186/gb-2014-15-6-r80
SVA retrotransposon insertion-associated deletion represents a novel mutational mechanism underlying large genomic copy number changes with non-recurrent breakpoints
Abstract
Background: Genomic disorders are caused by copy number changes that may exhibit recurrent breakpoints processed by nonallelic homologous recombination. However, region-specific disease-associated copy number changes have also been observed which exhibit non-recurrent breakpoints. The mechanisms underlying these non-recurrent copy number changes have not yet been fully elucidated.
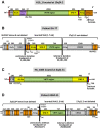
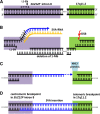
Results: We analyze large NF1 deletions with non-recurrent breakpoints as a model to investigate the full spectrum of causative mechanisms, and observe that they are mediated by various DNA double strand break repair mechanisms, as well as aberrant replication. Further, two of the 17 NF1 deletions with non-recurrent breakpoints, identified in unrelated patients, occur in association with the concomitant insertion of SINE/variable number of tandem repeats/Alu (SVA) retrotransposons at the deletion breakpoints. The respective breakpoints are refractory to analysis by standard breakpoint-spanning PCRs and are only identified by means of optimized PCR protocols designed to amplify across GC-rich sequences. The SVA elements are integrated within SUZ12P intron 8 in both patients, and were mediated by target-primed reverse transcription of SVA mRNA intermediates derived from retrotranspositionally active source elements. Both SVA insertions occurred during early postzygotic development and are uniquely associated with large deletions of 1 Mb and 867 kb, respectively, at the insertion sites.
Conclusions: Since active SVA elements are abundant in the human genome and the retrotranspositional activity of many SVA source elements is high, SVA insertion-associated large genomic deletions encompassing many hundreds of kilobases could constitute a novel and as yet under-appreciated mechanism underlying large-scale copy number changes in the human genome.
Figures



References
-
- Kluwe L, Siebert R, Gesk S, Friedrich RE, Tinschert S, Kehrer-Sawatzki H, Mautner V-F. Screening of 500 unselected neurofibromatosis 1 patients for deletions of the NF1 gene. Hum Mutat. 2004;23:111–116. - PubMed
-
- Kehrer-Sawatzki H, Cooper DN. In: Neurofibromatosis Type 1, Molecular and Cellular Biology. Upadhyaya M, Cooper DN, editor. Heidelberg: Springer; 2012. NF1 microdeletions and their underlying mutational mechanisms; pp. 187–211.
-
- Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K. NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet. 2000;9:35–46. - PubMed
-
- López-Correa C, Dorschner M, Brems H, Lazaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer-Sawatzki H, Rutkowski JL, Fryns JP, Marynen P, Stephens K, Legius E. Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet. 2001;10:1387–1392. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

