Cleavage by signal peptide peptidase is required for the degradation of selected tail-anchored proteins
- PMID: 24958774
- PMCID: PMC4068138
- DOI: 10.1083/jcb.201312009
Cleavage by signal peptide peptidase is required for the degradation of selected tail-anchored proteins
Abstract
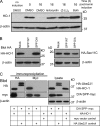
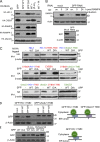
The regulated turnover of endoplasmic reticulum (ER)-resident membrane proteins requires their extraction from the membrane lipid bilayer and subsequent proteasome-mediated degradation. Cleavage within the transmembrane domain provides an attractive mechanism to facilitate protein dislocation but has never been shown for endogenous substrates. To determine whether intramembrane proteolysis, specifically cleavage by the intramembrane-cleaving aspartyl protease signal peptide peptidase (SPP), is involved in this pathway, we generated an SPP-specific somatic cell knockout. In a stable isotope labeling by amino acids in cell culture-based proteomics screen, we identified HO-1 (heme oxygenase-1), the rate-limiting enzyme in the degradation of heme to biliverdin, as a novel SPP substrate. Intramembrane cleavage by catalytically active SPP provided the primary proteolytic step required for the extraction and subsequent proteasome-dependent degradation of HO-1, an ER-resident tail-anchored protein. SPP-mediated proteolysis was not limited to HO-1 but was required for the dislocation and degradation of additional tail-anchored ER-resident proteins. Our study identifies tail-anchored proteins as novel SPP substrates and a specific requirement for SPP-mediated intramembrane cleavage in protein turnover.
© 2014 Boname et al.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

